An Anthropocentric View of the Virosphere-Host Relationship
- PMID: 28912772
- PMCID: PMC5582082
- DOI: 10.3389/fmicb.2017.01673
An Anthropocentric View of the Virosphere-Host Relationship
Abstract
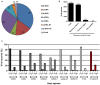
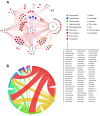
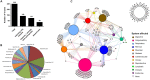
For over a century, viruses have been known as the most abundant and diverse group of organisms on Earth, forming a virosphere. Based on extensive meta-analyses, we present, for the first time, a wide and complete overview of virus-host network, covering all known viral species. Our data indicate that most of known viral species, regardless of their genomic category, have an intriguingly narrow host range, infecting only 1 or 2 host species. Our data also show that the known virosphere has expanded based on viruses of human interest, related to economical, medical or biotechnological activities. In addition, we provide an overview of the distribution of viruses on different environments on Earth, based on meta-analyses of available metaviromic data, showing the contrasting ubiquity of head-tailed phages against the specificity of some viral groups in certain environments. Finally, we uncovered all human viral species, exploring their diversity and the most affected organic systems. The virus-host network presented here shows an anthropocentric view of the virology. It is therefore clear that a huge effort and change in perspective is necessary to see more than the tip of the iceberg when it comes to virology.
Keywords: anthropocentric; metavirome; network; virosphere; virus–host relationship.
Figures

References
-
- Bastian M., Heymann S., Jacomy M. (2009). “Gephi: an open source software for exploring and manipulating networks,” in Third International AAAI Conference on Weblogs Social Media, Paris, 361–362. 10.1136/qshc.2004.010033 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

