Shared and Distinct Phenotypes and Functions of Human CD161++ Vα7.2+ T Cell Subsets
- PMID: 28912775
- PMCID: PMC5582200
- DOI: 10.3389/fimmu.2017.01031
Shared and Distinct Phenotypes and Functions of Human CD161++ Vα7.2+ T Cell Subsets
Abstract
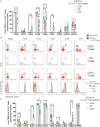
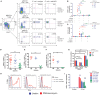
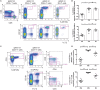
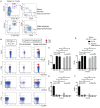
Human mucosal-associated invariant T (MAIT) cells are an important T cell subset that are enriched in tissues and possess potent effector functions. Typically such cells are marked by their expression of Vα7.2-Jα33/Jα20/Jα12 T cell receptors, and functionally they are major histocompatibility complex class I-related protein 1 (MR1)-restricted, responding to bacterially derived riboflavin synthesis intermediates. MAIT cells are contained within the CD161++ Vα7.2+ T cell population, the majority of which express the CD8 receptor (CD8+), while a smaller fraction expresses neither CD8 or CD4 coreceptor (double negative; DN) and a further minority are CD4+. Whether these cells have distinct homing patterns, phenotype and functions have not been examined in detail. We used a combination of phenotypic staining and functional assays to address the similarities and differences between these CD161++ Vα7.2+ T cell subsets. We find that most features are shared between CD8+ and DN CD161++ Vα7.2+ T cells, with a small but detectable role evident for CD8 binding in tuning functional responsiveness. By contrast, the CD4+ CD161++ Vα7.2+ T cell population, although showing MR1-dependent responsiveness to bacterial stimuli, display reduced T helper 1 effector functions, including cytolytic machinery, while retaining the capacity to secrete interleukin-4 (IL-4) and IL-13. This was consistent with underlying changes in transcription factor (TF) expression. Although we found that only a proportion of CD4+ CD161++ Vα7.2+ T cells stained for the MR1-tetramer, explaining some of the heterogeneity of CD4+ CD161++ Vα7.2+ T cells, these differences in TF expression were shared with CD4+ CD161++ MR1-tetramer+ cells. These data reveal the functional diversity of human CD161++ Vα7.2+ T cells and indicate potentially distinct roles for the different subsets in vivo.
Keywords: CD8 coreceptor; MHC class I-related protein 1; MHC class I-related protein 1-tetramer; innate-like T cells; mucosal-associated invariant T cells; subsets; transcription factors.
Figures





References
-
- Porcelli S, Yockey CE, Brenner MB, Balk SP. Analysis of T cell antigen receptor (TCR) expression by human peripheral blood CD4-8-alpha/beta T cells demonstrates preferential use of several V beta genes and an invariant TCR alpha chain. J Exp Med (1993) 178:1–16.10.1084/jem.178.1.1 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

