Haplotypes for Type, Degree, and Rate of Marbling in Cattle Are Syntenic with Human Muscular Dystrophy
- PMID: 28913347
- PMCID: PMC5585636
- DOI: 10.1155/2017/6532837
Haplotypes for Type, Degree, and Rate of Marbling in Cattle Are Syntenic with Human Muscular Dystrophy
Abstract
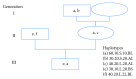
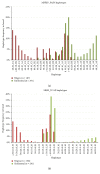
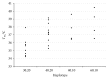
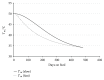
Traditional analyses of a QTL on Bota 19 implicate a surfeit of candidates, but each is of marginal significance in explaining the deposition of healthy, low melting temperature fat within marbled muscle of Wagyu cattle. As an alternative approach, we have used genomic, multigenerational segregation to identify 14 conserved, ancestral 20 Mb haplotypes. These determine the degree and rate of marbling in Wagyu and other breeds of cattle. The melting temperature of intramuscular fat is highly heritable and traceable by haplotyping. Fortunately, for the production of healthy beef, some of these haplotypes are sufficiently penetrant to be expressed in heterozygous crossbreds, thereby allowing selection of sires which will improve the healthiness of beef produced under even harsh climatic conditions. The region of Bota 19 is syntenic to a region of Hosa 17 known to be important in muscle metabolism and in determining susceptibility to a form of human muscular dystrophy.
Figures





References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

