Systems-Level Annotation of a Metabolomics Data Set Reduces 25 000 Features to Fewer than 1000 Unique Metabolites
- PMID: 28914531
- PMCID: PMC6427824
- DOI: 10.1021/acs.analchem.7b02380
Systems-Level Annotation of a Metabolomics Data Set Reduces 25 000 Features to Fewer than 1000 Unique Metabolites
Abstract
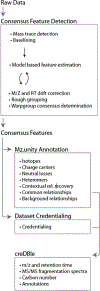
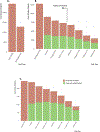
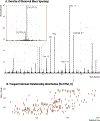
When using liquid chromatography/mass spectrometry (LC/MS) to perform untargeted metabolomics, it is now routine to detect tens of thousands of features from biological samples. Poor understanding of the data, however, has complicated interpretation and masked the number of unique metabolites actually being measured in an experiment. Here we place an upper bound on the number of unique metabolites detected in Escherichia coli samples analyzed with one untargeted metabolomics method. We first group multiple features arising from the same analyte, which we call "degenerate features", using a context-driven annotation approach. Surprisingly, this analysis revealed thousands of previously unreported degeneracies that reduced the number of unique analytes to ∼2961. We then applied an orthogonal approach to remove nonbiological features from the data using the 13C-based credentialing technology. This further reduced the number of unique analytes to less than 1000. Our 90% reduction in data is 5-fold greater than previously published studies. On the basis of the results, we propose an alternative approach to untargeted metabolomics that relies on thoroughly annotated reference data sets. To this end, we introduce the creDBle database ( http://creDBle.wustl.edu ), which contains accurate mass, retention time, and MS/MS fragmentation data as well as annotations of all credentialed features.
Figures

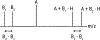
References
-
- Broeckling CD; Afsar FA; Neumann S; Ben-Hur A; Prenni JE Anal. Chem. 2014, 86 (14), 6812–6817. - PubMed
-
- Dunn WB; Erban A; Weber RJM; Creek DJ; Brown M; Breitling R; Hankemeier T; Goodacre R; Neumann S; Kopka J; Viant MR Metabolomics 2013, 9, 44–66.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

