Loss of p16INK4A stimulates aberrant mitochondrial biogenesis through a CDK4/Rb-independent pathway
- PMID: 28915557
- PMCID: PMC5593528
- DOI: 10.18632/oncotarget.19862
Loss of p16INK4A stimulates aberrant mitochondrial biogenesis through a CDK4/Rb-independent pathway
Abstract
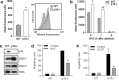
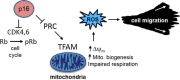
The tumor suppressor p16INK4A (p16) inhibits cell cycle progression through the CDK4/Rb pathway. We have previously shown that p16 regulates cellular oxidative stress, independent of its role in cell cycle control. We investigated whether loss of p16 had a direct impact on the mitochondria. We found that p16-null primary mouse fibroblasts (PMFs) displayed increased mitochondrial mass and expression of mitochondrial respiratory subunit proteins compared to wild-type (WT) PMFs. These findings in p16-null PMFs were associated with increased expression of the mitochondrial biogenesis transcription factors PRC and TFAM. On the other hand, p16-deficient PMFs demonstrated reduced mitochondrial respiration capacity consistent with electron microscopy findings showing that mitochondria in p16-deficient PMFs have abnormal morphology. Consistent with increased mitochondrial mass and reduced respiratory capacity, p16-deficient PMFs generated increased mitochondrial superoxide. One biological consequence of elevated ROS in p16-deficient PMFs was enhanced migration, which was reduced by the ROS scavenger N-acetylcysteine. Finally, p16-deficient PMFs displayed increased mitochondrial membrane potential, which was also required for their enhanced migration. The mitochondrial and migration phenotype was restored in p16-deficient PMFs by forced expression of p16. Similarly, over-expression of p16 in human melanocytes and A375 melanoma cells led to decreased expression of some mitochondrial respiratory proteins, enhanced respiration, and decreased migration. Inhibition of Rb phosphorylation in melanocytes and melanoma cells, either by addition of chemical CDK4 inhibitors or RNAi-mediated knockdown of CDK4, did not mimic the effects of p16 loss. These results suggest that p16 regulates mitochondrial biogenesis and function, which is independent of the canonical CDK4/Rb pathway.
Keywords: CDK4; fibroblast; migration; mitochondria; p16.
Conflict of interest statement
CONFLICTS OF INTEREST The authors declare that no conflicts of interest exist.
Figures






Similar articles
-
Altered mRNA expression of the Rb and p16 tumor suppressor genes and of CDK4 in transitional cell carcinomas of the urinary bladder associated with tumor progression.Anticancer Res. 2004 Mar-Apr;24(2B):1011-23. Anticancer Res. 2004. PMID: 15161057
-
Preimplantation-embryo-specific cell-cycle regulation is attributable to a low expression of retinoblastoma protein rather than its phosphorylation.J Reprod Dev. 2011 Sep;57(4):492-9. doi: 10.1262/jrd.10-170o. Epub 2011 Apr 26. J Reprod Dev. 2011. PMID: 21519154
-
The p16(INK4A) tumor suppressor regulates cellular oxidative stress.Oncogene. 2011 Jan 20;30(3):265-74. doi: 10.1038/onc.2010.419. Epub 2010 Sep 13. Oncogene. 2011. PMID: 20838381 Free PMC article.
-
p16INK4a-induced senescence is disabled by melanoma-associated mutations.Aging Cell. 2008 Oct;7(5):733-45. doi: 10.1111/j.1474-9726.2008.00422.x. Aging Cell. 2008. PMID: 18843795 Free PMC article.
-
The physiology of p16(INK4A)-mediated G1 proliferative arrest.Cell Biochem Biophys. 2000;33(2):189-97. doi: 10.1385/CBB:33:2:189. Cell Biochem Biophys. 2000. PMID: 11325039 Review.
Cited by
-
Bi-allelic Loss of CDKN2A Initiates Melanoma Invasion via BRN2 Activation.Cancer Cell. 2018 Jul 9;34(1):56-68.e9. doi: 10.1016/j.ccell.2018.05.014. Cancer Cell. 2018. PMID: 29990501 Free PMC article.
-
Endometrial Aging and Reproductive Decline: The Central Role of Mitochondrial Dysfunction.Int J Mol Sci. 2025 May 24;26(11):5060. doi: 10.3390/ijms26115060. Int J Mol Sci. 2025. PMID: 40507871 Free PMC article. Review.
-
A novel CDKN2A variant (p16L117P ) in a patient with familial and multiple primary melanomas.Pigment Cell Melanoma Res. 2019 Sep;32(5):734-738. doi: 10.1111/pcmr.12787. Epub 2019 May 3. Pigment Cell Melanoma Res. 2019. PMID: 31001908 Free PMC article.
-
Differential p16 expression levels in the liver, hepatocytes and hepatocellular cell lines.PeerJ. 2021 Nov 2;9:e12358. doi: 10.7717/peerj.12358. eCollection 2021. PeerJ. 2021. PMID: 34760375 Free PMC article.
-
Effects of the p16/cyclin D1/CDK4/Rb/E2F1 pathway on aberrant lung fibroblast proliferation in neonatal rats exposed to hyperoxia.Exp Ther Med. 2021 Oct;22(4):1057. doi: 10.3892/etm.2021.10491. Epub 2021 Jul 26. Exp Ther Med. 2021. PMID: 34434271 Free PMC article.
References
-
- Sharpless NE, DePinho RA. The INK4A/ARF locus and its two gene products. Curr Opin Genet Dev. 1999;9:22–30. - PubMed
-
- Zuo L, Weger J, Yang Q, Goldstein AM, Tucker MA, Walker GJ, Hayward N, Dracopoli NC. Germline mutations in the p16INK4a binding domain of CDK4 in familial melanoma. Nat Genet. 1996;12:97–99. - PubMed
-
- Hashemi J, Platz A, Ueno T, Stierner U, Ringborg U, Hansson J. CDKN2A germ-line mutations in individuals with multiple cutaneous melanomas. Cancer Res. 2000;60:6864–6867. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

