The cacao Criollo genome v2.0: an improved version of the genome for genetic and functional genomic studies
- PMID: 28915793
- PMCID: PMC5603072
- DOI: 10.1186/s12864-017-4120-9
The cacao Criollo genome v2.0: an improved version of the genome for genetic and functional genomic studies
Abstract
Background: Theobroma cacao L., native to the Amazonian basin of South America, is an economically important fruit tree crop for tropical countries as a source of chocolate. The first draft genome of the species, from a Criollo cultivar, was published in 2011. Although a useful resource, some improvements are possible, including identifying misassemblies, reducing the number of scaffolds and gaps, and anchoring un-anchored sequences to the 10 chromosomes.
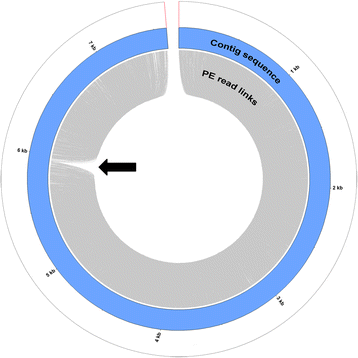
Methods: We used a NGS-based approach to significantly improve the assembly of the Belizian Criollo B97-61/B2 genome. We combined four Illumina large insert size mate paired libraries with 52x of Pacific Biosciences long reads to correct misassembled regions and reduced the number of scaffolds. We then used genotyping by sequencing (GBS) methods to increase the proportion of the assembly anchored to chromosomes.
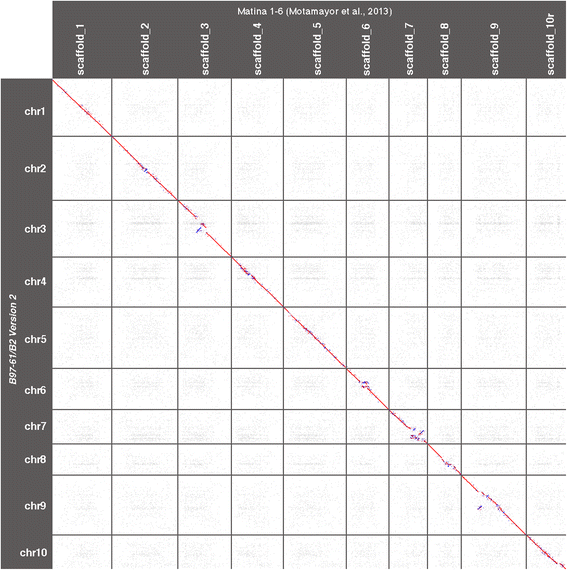
Results: The scaffold number decreased from 4,792 in assembly V1 to 554 in V2 while the scaffold N50 size has increased from 0.47 Mb in V1 to 6.5 Mb in V2. A total of 96.7% of the assembly was anchored to the 10 chromosomes compared to 66.8% in the previous version. Unknown sites (Ns) were reduced from 10.8% to 5.7%. In addition, we updated the functional annotations and performed a new RefSeq structural annotation based on RNAseq evidence.
Conclusion: Theobroma cacao Criollo genome version 2 will be a valuable resource for the investigation of complex traits at the genomic level and for future comparative genomics and genetics studies in cacao tree. New functional tools and annotations are available on the Cocoa Genome Hub ( http://cocoa-genome-hub.southgreen.fr ).
Keywords: Criollo B97–61/B2 genome; GBS; Genome Assembly; Mate Paired sequences; Theobroma cacao.
Conflict of interest statement
Ethics approval and consent to participate
The
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
-
- Argout X, Fouet O, Wincker P, Gramacho K, Legavre T, Sabau X, et al. Towards the understanding of the cocoa transcriptome: Production and analysis of an exhaustive dataset of ESTs of Theobroma cacao L. generated from various tissues and under various conditions. BMC Genomics. 2008;9:512. - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

