Replicability of time-varying connectivity patterns in large resting state fMRI samples
- PMID: 28916181
- PMCID: PMC5775892
- DOI: 10.1016/j.neuroimage.2017.09.020
Replicability of time-varying connectivity patterns in large resting state fMRI samples
Abstract
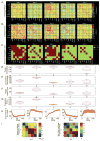
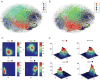
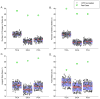
The past few years have seen an emergence of approaches that leverage temporal changes in whole-brain patterns of functional connectivity (the chronnectome). In this chronnectome study, we investigate the replicability of the human brain's inter-regional coupling dynamics during rest by evaluating two different dynamic functional network connectivity (dFNC) analysis frameworks using 7 500 functional magnetic resonance imaging (fMRI) datasets. To quantify the extent to which the emergent functional connectivity (FC) patterns are reproducible, we characterize the temporal dynamics by deriving several summary measures across multiple large, independent age-matched samples. Reproducibility was demonstrated through the existence of basic connectivity patterns (FC states) amidst an ensemble of inter-regional connections. Furthermore, application of the methods to conservatively configured (statistically stationary, linear and Gaussian) surrogate datasets revealed that some of the studied state summary measures were indeed statistically significant and also suggested that this class of null model did not explain the fMRI data fully. This extensive testing of reproducibility of similarity statistics also suggests that the estimated FC states are robust against variation in data quality, analysis, grouping, and decomposition methods. We conclude that future investigations probing the functional and neurophysiological relevance of time-varying connectivity assume critical importance.
Keywords: Dynamic functional connectivity; Fuzzy meta-state; Hard clustering; Replicability; Sliding window method; Surrogate testing.
Copyright © 2017 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing financial interests.
Figures







Similar articles
-
Resting state networks in empirical and simulated dynamic functional connectivity.Neuroimage. 2017 Oct 1;159:388-402. doi: 10.1016/j.neuroimage.2017.07.065. Epub 2017 Aug 3. Neuroimage. 2017. PMID: 28782678
-
The chronnectome: Evaluating replicability of dynamic connectivity patterns in 7500 resting fMRI datasets.Annu Int Conf IEEE Eng Med Biol Soc. 2016 Aug;2016:5571-5574. doi: 10.1109/EMBC.2016.7591989. Annu Int Conf IEEE Eng Med Biol Soc. 2016. PMID: 28269517
-
Characterizing dynamic amplitude of low-frequency fluctuation and its relationship with dynamic functional connectivity: An application to schizophrenia.Neuroimage. 2018 Oct 15;180(Pt B):619-631. doi: 10.1016/j.neuroimage.2017.09.035. Epub 2017 Sep 20. Neuroimage. 2018. PMID: 28939432 Free PMC article.
-
Interpreting temporal fluctuations in resting-state functional connectivity MRI.Neuroimage. 2017 Dec;163:437-455. doi: 10.1016/j.neuroimage.2017.09.012. Epub 2017 Sep 12. Neuroimage. 2017. PMID: 28916180 Review.
-
Mapping cognitive and emotional networks in neurosurgical patients using resting-state functional magnetic resonance imaging.Neurosurg Focus. 2020 Feb 1;48(2):E9. doi: 10.3171/2019.11.FOCUS19773. Neurosurg Focus. 2020. PMID: 32006946 Free PMC article. Review.
Cited by
-
NeuroMark: An automated and adaptive ICA based pipeline to identify reproducible fMRI markers of brain disorders.Neuroimage Clin. 2020;28:102375. doi: 10.1016/j.nicl.2020.102375. Epub 2020 Aug 11. Neuroimage Clin. 2020. PMID: 32961402 Free PMC article.
-
Data-driven approaches for identifying links between brain structure and function in health and disease.Dialogues Clin Neurosci. 2018 Jun;20(2):87-99. doi: 10.31887/DCNS.2018.20.2/vcalhoun. Dialogues Clin Neurosci. 2018. PMID: 30250386 Free PMC article.
-
Disrupted dynamic functional network connectivity in fetal alcohol spectrum disorders.Alcohol Clin Exp Res (Hoboken). 2023 Apr;47(4):687-703. doi: 10.1111/acer.15046. Epub 2023 Mar 15. Alcohol Clin Exp Res (Hoboken). 2023. PMID: 36880528 Free PMC article.
-
Reduced higher dimensional temporal dynamism in neurofibromatosis type 1.Neuroimage Clin. 2019;22:101692. doi: 10.1016/j.nicl.2019.101692. Epub 2019 Jan 29. Neuroimage Clin. 2019. PMID: 30710873 Free PMC article.
-
Resting State Dynamic Functional Connectivity in Neurodegenerative Conditions: A Review of Magnetic Resonance Imaging Findings.Front Neurosci. 2019 Jun 20;13:657. doi: 10.3389/fnins.2019.00657. eCollection 2019. Front Neurosci. 2019. PMID: 31281241 Free PMC article. Review.
References
-
- Abrol A, Chaze C, Damaraju E, Calhoun VD. The chronnectome: replicability of dynamic connectivity patterns in 7500 resting fmri datasets. IEEE Engineering in Medicine and Biology Society. 2016:5571–5574. - PubMed
-
- Allen EA, Erhardt EB, Damaraju E, Gruner W, Segall JM, Silva RF, Havlicek M, Rachakonda S, Fries J, Kalyanam R, Michael AM, Caprihan A, Turner JA, Eichele T, Adelsheim S, Bryan AD, Bustillo J, Clark VP, Feldstein Ewing SW, Filbey F, Ford CC, Hutchison K, Jung RE, Kiehl KA, Kodituwakku P, Komesu YM, Mayer AR, Pearlson GD, Phillips JP, Sadek JR, Stevens M, Teuscher U, Thoma RJ, Calhoun VD. A baseline for the multivariate comparison of resting state networks. Front Syst Neurosci. 2011;5 - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

