Glycans Function as Anchors for Antibodies and Help Drive HIV Broadly Neutralizing Antibody Development
- PMID: 28916265
- PMCID: PMC5613947
- DOI: 10.1016/j.immuni.2017.08.006
Glycans Function as Anchors for Antibodies and Help Drive HIV Broadly Neutralizing Antibody Development
Erratum in
-
Glycans Function as Anchors for Antibodies and Help Drive HIV Broadly Neutralizing Antibody Development.Immunity. 2017 Nov 21;47(5):1004. doi: 10.1016/j.immuni.2017.10.012. Immunity. 2017. PMID: 29166578 Free PMC article. No abstract available.
Abstract
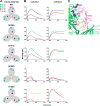
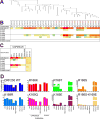
Apex broadly neutralizing HIV antibodies (bnAbs) recognize glycans and protein surface close to the 3-fold axis of the envelope (Env) trimer and are among the most potent and broad Abs described. The evolution of apex bnAbs from one donor (CAP256) has been studied in detail and many Abs at different stages of maturation have been described. Using diverse engineering tools, we investigated the involvement of glycan recognition in the development of the CAP256.VRC26 Ab lineage. We found that sialic acid-bearing glycans were recognized by germline-encoded and somatically mutated residues on the Ab heavy chain. This recognition provided an "anchor" for the Abs as the core protein epitope varies, prevented complete neutralization escape, and eventually led to broadening of the response. These findings illustrate how glycan-specific maturation enables a human Ab to cope with pathogen escape mechanisms and will aid in optimization of immunization strategies to induce V2 apex bnAb responses.
Keywords: B cell affinity maturation; Env glycans; HIV envelope trimer; V2 apex epitope; bnAbs; broadly neutralizing antibodies; glycan engineering; virus escape.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures




References
-
- Bonsignori M, Hwang KK, Chen X, Tsao CY, Morris L, Gray E, Marshall DJ, Crump JA, Kapiga SH, Sam NE, et al. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. Journal of virology. 2011;85:9998–10009. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

