DELLA-GAF1 Complex Is a Main Component in Gibberellin Feedback Regulation of GA20 Oxidase 2
- PMID: 28916594
- PMCID: PMC5664458
- DOI: 10.1104/pp.17.00282
DELLA-GAF1 Complex Is a Main Component in Gibberellin Feedback Regulation of GA20 Oxidase 2
Abstract
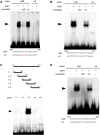
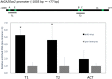
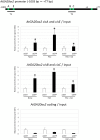
Gibberellins (GAs) are phytohormones that regulate many aspects of plant growth and development, including germination, elongation, flowering, and floral development. Negative feedback regulation contributes to homeostasis of the GA level. DELLAs are negative regulators of GA signaling and are rapidly degraded in the presence of GAs. DELLAs regulate many target genes, including AtGA20ox2 in Arabidopsis (Arabidopsis thaliana), encoding the GA-biosynthetic enzyme GA 20-oxidase. As DELLAs do not have an apparent DNA-binding motif, transcription factors that act in association with DELLA are necessary for regulating the target genes. Previous studies have identified GAI-ASSOCIATED FACTOR1 (GAF1) as such a DELLA interactor, with which DELLAs act as coactivators, and AtGA20ox2 was identified as a target gene of the DELLA-GAF1 complex. In this study, electrophoretic mobility shift and chromatin immunoprecipitation assays showed that four GAF1-binding sites exist in the AtGA20ox2 promoter. Using transgenic plants, we further evaluated the contribution of the DELLA-GAF1 complex to GA feedback regulation. Mutations in four GAF1-binding sites abolished the negative feedback of AtGA20ox2 in transgenic plants. Our results showed that GAF1-binding sites are necessary for GA feedback regulation of AtGA20ox2, suggesting that the DELLA-GAF1 complex is a main component of the GA feedback regulation of AtGA20ox2.
© 2017 American Society of Plant Biologists. All Rights Reserved.
Figures





Similar articles
-
DELLA-GAF1 complex is involved in tissue-specific expression and gibberellin feedback regulation of GA20ox1 in Arabidopsis.Plant Mol Biol. 2021 Oct;107(3):147-158. doi: 10.1007/s11103-021-01195-z. Epub 2021 Sep 25. Plant Mol Biol. 2021. PMID: 34562198
-
Binding of GID1 to DELLAs promotes dissociation of GAF1 from DELLA in GA dependent manner.Plant Signal Behav. 2015;10(10):e1052923. doi: 10.1080/15592324.2015.1052923. Plant Signal Behav. 2015. PMID: 26237582 Free PMC article.
-
DELLAs function as coactivators of GAI-ASSOCIATED FACTOR1 in regulation of gibberellin homeostasis and signaling in Arabidopsis.Plant Cell. 2014 Jul;26(7):2920-38. doi: 10.1105/tpc.114.125690. Epub 2014 Jul 17. Plant Cell. 2014. PMID: 25035403 Free PMC article.
-
DELLA and SCL3 balance gibberellin feedback regulation by utilizing INDETERMINATE DOMAIN proteins as transcriptional scaffolds.Plant Signal Behav. 2014;9(9):e29726. doi: 10.4161/psb.29726. Plant Signal Behav. 2014. PMID: 25763707 Free PMC article. Review.
-
Molecular biology of gibberellins signaling in higher plants.Int Rev Cell Mol Biol. 2008;268:191-221. doi: 10.1016/S1937-6448(08)00806-X. Int Rev Cell Mol Biol. 2008. PMID: 18703407 Review.
Cited by
-
CRISPR/Cas9 Directed Mutagenesis of OsGA20ox2 in High Yielding Basmati Rice (Oryza sativa L.) Line and Comparative Proteome Profiling of Unveiled Changes Triggered by Mutations.Int J Mol Sci. 2020 Aug 26;21(17):6170. doi: 10.3390/ijms21176170. Int J Mol Sci. 2020. PMID: 32859098 Free PMC article.
-
A LBD transcription factor from moso bamboo, PheLBD12, regulates plant height in transgenic rice.Plant Mol Biol. 2024 Sep 3;114(5):95. doi: 10.1007/s11103-024-01487-0. Plant Mol Biol. 2024. PMID: 39223419
-
Roles of Abscisic Acid and Gibberellins in Stem/Root Tuber Development.Int J Mol Sci. 2022 Apr 29;23(9):4955. doi: 10.3390/ijms23094955. Int J Mol Sci. 2022. PMID: 35563355 Free PMC article. Review.
-
GROWTH-REGULATING FACTORS Interact with DELLAs and Regulate Growth in Cold Stress.Plant Cell. 2020 Apr;32(4):1018-1034. doi: 10.1105/tpc.19.00784. Epub 2020 Feb 14. Plant Cell. 2020. PMID: 32060178 Free PMC article.
-
The Roles of Mepiquate Chloride and Melatonin in the Morpho-Physiological Activity of Cotton under Abiotic Stress.Int J Mol Sci. 2023 Dec 23;25(1):235. doi: 10.3390/ijms25010235. Int J Mol Sci. 2023. PMID: 38203405 Free PMC article. Review.
References
-
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 - PubMed
-
- Colasanti J, Yuan Z, Sundaresan V (1998) The indeterminate gene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize. Cell 93: 593–603 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

