Complete fusion of a transposon and herpesvirus created the Teratorn mobile element in medaka fish
- PMID: 28916771
- PMCID: PMC5601938
- DOI: 10.1038/s41467-017-00527-2
Complete fusion of a transposon and herpesvirus created the Teratorn mobile element in medaka fish
Abstract
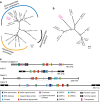
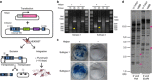
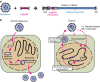
Mobile genetic elements (e.g., transposable elements and viruses) display significant diversity with various life cycles, but how novel elements emerge remains obscure. Here, we report a giant (180-kb long) transposon, Teratorn, originally identified in the genome of medaka, Oryzias latipes. Teratorn belongs to the piggyBac superfamily and retains the transposition activity. Remarkably, Teratorn is largely derived from a herpesvirus of the Alloherpesviridae family that could infect fish and amphibians. Genomic survey of Teratorn-like elements reveals that some of them exist as a fused form between piggyBac transposon and herpesvirus genome in teleosts, implying the generality of transposon-herpesvirus fusion. We propose that Teratorn was created by a unique fusion of DNA transposon and herpesvirus, leading to life cycle shift. Our study supports the idea that recombination is the key event in generation of novel mobile genetic elements. Teratorn is a large mobile genetic element originally identified in the small teleost fish medaka. Here, the authors show that Teratorn is derived from the fusion of a piggyBac superfamily DNA transposon and an alloherpesvirus and that it is widely found across teleost fish.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



Similar articles
-
Teratorn and its relatives - a cross-point of distinct mobile elements, transposons and viruses.Front Vet Sci. 2023 Apr 28;10:1158023. doi: 10.3389/fvets.2023.1158023. eCollection 2023. Front Vet Sci. 2023. PMID: 37187934 Free PMC article. Review.
-
Fusion of piggyBac-like transposons and herpesviruses occurs frequently in teleosts.Zoological Lett. 2018 Feb 21;4:6. doi: 10.1186/s40851-018-0089-8. eCollection 2018. Zoological Lett. 2018. PMID: 29484202 Free PMC article.
-
Teratorn and Its Related Elements - a Novel Group of Herpesviruses Widespread in Teleost Genomes.Zoolog Sci. 2023 Apr;40(2):83-90. doi: 10.2108/zs220069. Zoolog Sci. 2023. PMID: 37042688 Review.
-
Transposable elements in medaka fish.Zoolog Sci. 2002 Jan;19(1):1-6. doi: 10.2108/zsj.19.1. Zoolog Sci. 2002. PMID: 12025396 Review.
-
Excision of the tol2 transposable element of the medaka fish, Oryzias latipes, in zebrafish, Danio rerio.Gene. 1998 Dec 28;225(1-2):17-22. doi: 10.1016/s0378-1119(98)00537-x. Gene. 1998. PMID: 9931412
Cited by
-
From parasites to partners: exploring the intricacies of host-transposon dynamics and coevolution.Funct Integr Genomics. 2023 Aug 23;23(3):278. doi: 10.1007/s10142-023-01206-w. Funct Integr Genomics. 2023. PMID: 37610667 Review.
-
Viruses Defined by the Position of the Virosphere within the Replicator Space.Microbiol Mol Biol Rev. 2021 Dec 15;85(4):e0019320. doi: 10.1128/MMBR.00193-20. Epub 2021 Sep 1. Microbiol Mol Biol Rev. 2021. PMID: 34468181 Free PMC article.
-
Virus-Host Coevolution with a Focus on Animal and Human DNA Viruses.J Mol Evol. 2020 Jan;88(1):41-56. doi: 10.1007/s00239-019-09913-4. Epub 2019 Oct 10. J Mol Evol. 2020. PMID: 31599342 Free PMC article. Review.
-
The Enterprise, a massive transposon carrying Spok meiotic drive genes.Genome Res. 2021 May;31(5):789-798. doi: 10.1101/gr.267609.120. Epub 2021 Apr 19. Genome Res. 2021. PMID: 33875482 Free PMC article.
-
Giant Starship Elements Mobilize Accessory Genes in Fungal Genomes.Mol Biol Evol. 2022 May 3;39(5):msac109. doi: 10.1093/molbev/msac109. Mol Biol Evol. 2022. PMID: 35588244 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

