Functional organization of cytoplasmic inclusion bodies in cells infected by respiratory syncytial virus
- PMID: 28916773
- PMCID: PMC5601476
- DOI: 10.1038/s41467-017-00655-9
Functional organization of cytoplasmic inclusion bodies in cells infected by respiratory syncytial virus
Abstract
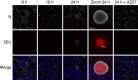
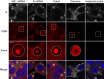
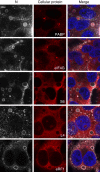
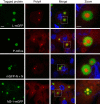
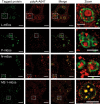
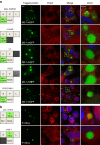
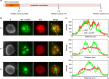
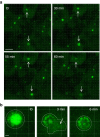
Infection of cells by respiratory syncytial virus induces the formation of cytoplasmic inclusion bodies (IBs) where all the components of the viral RNA polymerase complex are concentrated. However, the exact organization and function of these IBs remain unclear. In this study, we use conventional and super-resolution imaging to dissect the internal structure of IBs. We observe that newly synthetized viral mRNA and the viral transcription anti-terminator M2-1 concentrate in IB sub-compartments, which we term "IB-associated granules" (IBAGs). In contrast, viral genomic RNA, the nucleoprotein, the L polymerase and its cofactor P are excluded from IBAGs. Live imaging reveals that IBAGs are highly dynamic structures. Our data show that IBs are the main site of viral RNA synthesis. They further suggest that shortly after synthesis in IBs, viral mRNAs and M2-1 transiently concentrate in IBAGs before reaching the cytosol and suggest a novel post-transcriptional function for M2-1.Respiratory syncytial virus (RSV) induces formation of inclusion bodies (IBs) sheltering viral RNA synthesis. Here, Rincheval et al. identify highly dynamic IB-associated granules (IBAGs) that accumulate newly synthetized viral mRNA and the viral M2-1 protein but exclude viral genomic RNA and RNA polymerase complexes.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

