The mechano-sensing role of the unique SH3 insertion in plakin domains revealed by Molecular Dynamics simulations
- PMID: 28916774
- PMCID: PMC5601466
- DOI: 10.1038/s41598-017-11017-2
The mechano-sensing role of the unique SH3 insertion in plakin domains revealed by Molecular Dynamics simulations
Abstract
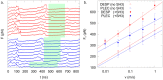
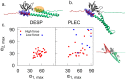
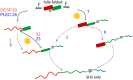
The plakin family of proteins, important actors in cross-linking force-bearing structures in the cell, contain a curious SH3 domain insertion in their chain of spectrin repeats (SRs). While SH3 domains are known to mediate protein-protein interactions, here, its canonical binding site is autoinhibited by the preceding SR. Under force, however, this SH3 domain could be released, and possibly launch a signaling cascade. We performed large-scale force-probe molecular dynamics simulations, across two orders of magnitude of loading rates, to test this hypothesis, on two prominent members of the plakin family: desmoplakin and plectin, obligate proteins at desmosomes and hemidesmosomes, respectively. Our simulations show that force unravels the SRs and abolishes the autoinhibition of the SH3 domain, an event well separated from the unfolding of this domain. The SH3 domain is free and fully functional for a significant portion of the unfolding trajectories. The rupture forces required for the two proteins significantly decrease when the SH3 domain is removed, which implies that the SH3 domain also stabilizes this junction. Our results persist across all simulations, and support a force-sensing as well as a stabilizing role of the unique SH3 insertion, putting forward this protein family as a new class of mechano-sensors.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
The structure of the plakin domain of plectin reveals a non-canonical SH3 domain interacting with its fourth spectrin repeat.J Biol Chem. 2011 Apr 8;286(14):12429-38. doi: 10.1074/jbc.M110.197467. Epub 2011 Feb 1. J Biol Chem. 2011. PMID: 21288893 Free PMC article.
-
Crystal structure of a rigid four-spectrin-repeat fragment of the human desmoplakin plakin domain.J Mol Biol. 2011 Jun 24;409(5):800-12. doi: 10.1016/j.jmb.2011.04.046. Epub 2011 Apr 22. J Mol Biol. 2011. PMID: 21536047 Free PMC article.
-
The Structure of the Plakin Domain of Plectin Reveals an Extended Rod-like Shape.J Biol Chem. 2016 Sep 2;291(36):18643-62. doi: 10.1074/jbc.M116.732909. Epub 2016 Jul 13. J Biol Chem. 2016. PMID: 27413182 Free PMC article.
-
The plakin domain of C. elegans VAB-10/plectin acts as a hub in a mechanotransduction pathway to promote morphogenesis.Development. 2019 Dec 13;146(24):dev183780. doi: 10.1242/dev.183780. Development. 2019. PMID: 31784459 Free PMC article.
-
Plakins in development and disease.Exp Cell Res. 2007 Jun 10;313(10):2189-203. doi: 10.1016/j.yexcr.2007.03.039. Epub 2007 Apr 19. Exp Cell Res. 2007. PMID: 17499243 Review.
Cited by
-
Biomaterial-based mechanical regulation facilitates scarless wound healing with functional skin appendage regeneration.Mil Med Res. 2024 Feb 18;11(1):13. doi: 10.1186/s40779-024-00519-6. Mil Med Res. 2024. PMID: 38369464 Free PMC article. Review.
-
Scaling up single-cell mechanics to multicellular tissues - the role of the intermediate filament-desmosome network.J Cell Sci. 2020 Mar 16;133(6):jcs228031. doi: 10.1242/jcs.228031. J Cell Sci. 2020. PMID: 32179593 Free PMC article. Review.
-
Gustavson syndrome is caused by an in-frame deletion in RBMX associated with potentially disturbed SH3 domain interactions.Eur J Hum Genet. 2024 Mar;32(3):333-341. doi: 10.1038/s41431-023-01392-y. Epub 2023 Jun 5. Eur J Hum Genet. 2024. PMID: 37277488 Free PMC article.
-
How cytoskeletal crosstalk makes cells move: Bridging cell-free and cell studies.Biophys Rev (Melville). 2024 Jun 3;5(2):021307. doi: 10.1063/5.0198119. eCollection 2024 Jun. Biophys Rev (Melville). 2024. PMID: 38840976 Free PMC article. Review.
-
Intermediate filaments and their associated molecules.J Biomed Res. 2025 Feb 8;39(3):242-253. doi: 10.7555/JBR.38.20240193. J Biomed Res. 2025. PMID: 39930669 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

