The muscle regulatory transcription factor MyoD participates with p53 to directly increase the expression of the pro-apoptotic Bcl2 family member PUMA
- PMID: 28918507
- PMCID: PMC5693709
- DOI: 10.1007/s10495-017-1423-x
The muscle regulatory transcription factor MyoD participates with p53 to directly increase the expression of the pro-apoptotic Bcl2 family member PUMA
Abstract
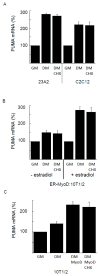
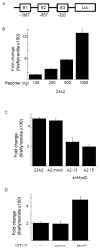
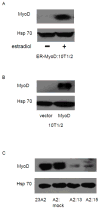
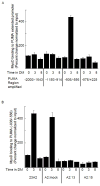
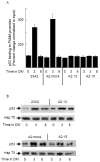
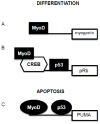
The muscle regulatory transcription factor MyoD is a master regulator of skeletal myoblast differentiation. We have previously reported that MyoD is also necessary for the elevated expression of the pro-apoptotic Bcl2 family member PUMA, and the ensuing apoptosis, that occurs in a subset of myoblasts induced to differentiate. Herein, we report the identification of a functional MyoD binding site within the extended PUMA promoter. In silico analysis of the murine PUMA extended promoter revealed three potential MyoD binding sites within 2 kb of the transcription start site. Expression from a luciferase reporter construct containing this 2 kb fragment was enhanced by activation of MyoD in both myoblasts and fibroblasts and diminished by silencing of MyoD in myoblasts. Experiments utilizing truncated versions of this promoter region revealed that the potential binding site at position - 857 was necessary for expression. Chromatin immunoprecipitation (ChIP) analysis confirmed binding of MyoD to the DNA region encompassing position - 857. The increase in MyoD binding to the PUMA promoter as a consequence of culture in differentiation media (DM) was comparable to the increase in MyoD binding at the myogenin promoter and was diminished in myoblasts silenced for MyoD expression. Finally, ChIP analysis using an antibody specific for the transcription factor p53 demonstrated that, in myoblasts silenced for MyoD expression, p53 binding to the PUMA promoter was diminished in response to culture in DM. These data indicate that MyoD plays a direct role in regulating PUMA expression and reveal functional consequences of MyoD expression on p53 mediated transcription of PUMA.
Keywords: Apoptosis; MyoD; P53; PUMA.
Figures


Similar articles
-
Increased expression of the pro-apoptotic Bcl2 family member PUMA and apoptosis by the muscle regulatory transcription factor MyoD in response to a variety of stimuli.Apoptosis. 2010 Jan;15(1):71-82. doi: 10.1007/s10495-009-0428-5. Apoptosis. 2010. PMID: 19943111
-
Increased expression of the pro-apoptotic Bcl2 family member PUMA is required for mitochondrial release of cytochrome C and the apoptosis associated with skeletal myoblast differentiation.Apoptosis. 2007 Dec;12(12):2143-54. doi: 10.1007/s10495-007-0135-z. Apoptosis. 2007. PMID: 17879164
-
p38α MAPK disables KMT1A-mediated repression of myogenic differentiation program.Skelet Muscle. 2016 Aug 22;6:28. doi: 10.1186/s13395-016-0100-z. eCollection 2016. Skelet Muscle. 2016. PMID: 27551368 Free PMC article.
-
Regulating a master regulator: establishing tissue-specific gene expression in skeletal muscle.Epigenetics. 2010 Nov-Dec;5(8):691-5. doi: 10.4161/epi.5.8.13045. Epub 2010 Nov 1. Epigenetics. 2010. PMID: 20716948 Free PMC article. Review.
-
Master control: transcriptional regulation of mammalian Myod.J Muscle Res Cell Motil. 2019 Jun;40(2):211-226. doi: 10.1007/s10974-019-09538-6. Epub 2019 Jul 12. J Muscle Res Cell Motil. 2019. PMID: 31301002 Free PMC article. Review.
Cited by
-
A structure-guided molecular chaperone approach for restoring the transcriptional activity of the p53 cancer mutant Y220C.Future Med Chem. 2019 Oct;11(19):2491-2504. doi: 10.4155/fmc-2019-0181. Future Med Chem. 2019. PMID: 31633398 Free PMC article.
-
Identification of the Myogenetic Oligodeoxynucleotides (myoDNs) That Promote Differentiation of Skeletal Muscle Myoblasts by Targeting Nucleolin.Front Cell Dev Biol. 2021 Jan 11;8:616706. doi: 10.3389/fcell.2020.616706. eCollection 2020. Front Cell Dev Biol. 2021. PMID: 33585451 Free PMC article.
-
Marek's Disease Virus (Gallid alphaherpesvirus 2)-Encoded miR-M2-5p Simultaneously Promotes Cell Proliferation and Suppresses Apoptosis Through RBM24 and MYOD1-Mediated Signaling Pathways.Front Microbiol. 2020 Nov 3;11:596422. doi: 10.3389/fmicb.2020.596422. eCollection 2020. Front Microbiol. 2020. PMID: 33224130 Free PMC article.
-
Development of the 12-Base Short Dimeric Myogenetic Oligodeoxynucleotide That Induces Myogenic Differentiation.BioTech (Basel). 2024 Apr 25;13(2):11. doi: 10.3390/biotech13020011. BioTech (Basel). 2024. PMID: 38804293 Free PMC article.
-
Mitochondrial stress response and myogenic differentiation.Front Cell Dev Biol. 2024 Apr 12;12:1381417. doi: 10.3389/fcell.2024.1381417. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 38681520 Free PMC article. Review.
References
-
- Ten Broek RW, Grefte S, Von den Hoff JW. Regulatory factors and cell populations involved in skeletal muscle regeneration. J Cell Physiol. 2010;224(1):7–16. - PubMed
-
- Partridge TA, Morgan JE. Multiple insights from myogenic cell transplants. Hum Gene Ther. 2014;25(5):404–5. - PubMed
-
- Grenier G, Rudnicki MA. The potential use of myogenic stem cells in regenerative medicine. Handb Exp Pharmacol. 2006;174:299–317. - PubMed
-
- Skuk D, Caron NJ, Goulet M, Roy B, Tremblay JP. Resetting the problem of cell death following muscle-derived cell transplantation: detection, dynamics and mechanisms. J Neuropathol Exp Neurol. 2003;62:951–67. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

