SAGA Is a General Cofactor for RNA Polymerase II Transcription
- PMID: 28918903
- PMCID: PMC5632562
- DOI: 10.1016/j.molcel.2017.08.016
SAGA Is a General Cofactor for RNA Polymerase II Transcription
Erratum in
-
SAGA Is a General Cofactor for RNA Polymerase II Transcription.Mol Cell. 2018 Jun 21;70(6):1163-1164. doi: 10.1016/j.molcel.2018.06.007. Mol Cell. 2018. PMID: 29932906 No abstract available.
Abstract
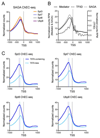
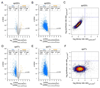
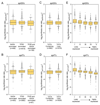
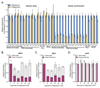
Prior studies suggested that SAGA and TFIID are alternative factors that promote RNA polymerase II transcription, with about 10% of genes in S. cerevisiae dependent on SAGA. We reassessed the role of SAGA by mapping its genome-wide location and role in global transcription in budding yeast. We find that SAGA maps to the UAS elements of most genes, overlapping with Mediator binding and irrespective of previous designations of SAGA- or TFIID-dominated genes. Disruption of SAGA through mutation or rapid subunit depletion reduces transcription from nearly all genes, measured by newly synthesized RNA. We also find that the acetyltransferase Gcn5 synergizes with Spt3 to promote global transcription and that Spt3 functions to stimulate TBP recruitment at all tested genes. Our data demonstrate that SAGA acts as a general cofactor required for essentially all RNA polymerase II transcription and is not consistent with the previous classification of SAGA- and TFIID-dominated genes.
Keywords: RNA polymerase II; SAGA complex; TATA box; TFIID complex; coactivator; transcription initiation.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures

References
-
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. - PubMed
-
- Basehoar AD, Zanton SJ, Pugh BF. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116:699–709. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

