Metabolic programming of the epigenome: host and gut microbial metabolite interactions with host chromatin
- PMID: 28919341
- PMCID: PMC5659875
- DOI: 10.1016/j.trsl.2017.08.005
Metabolic programming of the epigenome: host and gut microbial metabolite interactions with host chromatin
Abstract
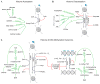
The mammalian gut microbiota has been linked to host developmental, immunologic, and metabolic outcomes. This collection of trillions of microbes inhabits the gut and produces a myriad of metabolites, which are measurable in host circulation and contribute to the pathogenesis of human diseases. The link between endogenous metabolite availability and chromatin regulation is a well-established and active area of investigation; however, whether microbial metabolites can elicit similar effects is less understood. In this review, we focus on seminal and recent research that establishes chromatin regulatory roles for both endogenous and microbial metabolites. We also highlight key physiologic and disease settings where microbial metabolite-host chromatin interactions have been established and/or may be pertinent.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures




Similar articles
-
The Gut Microbiota and their Metabolites: Potential Implications for the Host Epigenome.Adv Exp Med Biol. 2016;902:33-44. doi: 10.1007/978-3-319-31248-4_3. Adv Exp Med Biol. 2016. PMID: 27161349 Review.
-
Chromatin dynamics and histone modifications in intestinal microbiota-host crosstalk.Mol Metab. 2020 Aug;38:100925. doi: 10.1016/j.molmet.2019.12.005. Epub 2019 Dec 27. Mol Metab. 2020. PMID: 31992511 Free PMC article. Review.
-
Chemical signaling between gut microbiota and host chromatin: What is your gut really saying?J Biol Chem. 2017 May 26;292(21):8582-8593. doi: 10.1074/jbc.R116.761577. Epub 2017 Apr 7. J Biol Chem. 2017. PMID: 28389558 Free PMC article. Review.
-
Deciphering the Chemical Lexicon of Host-Gut Microbiota Interactions.Trends Pharmacol Sci. 2019 Jun;40(6):430-445. doi: 10.1016/j.tips.2019.04.006. Epub 2019 May 9. Trends Pharmacol Sci. 2019. PMID: 31079848 Free PMC article. Review.
-
Chemical Mechanisms of Colonization Resistance by the Gut Microbial Metabolome.ACS Chem Biol. 2020 May 15;15(5):1119-1126. doi: 10.1021/acschembio.9b00813. Epub 2020 Jan 14. ACS Chem Biol. 2020. PMID: 31895538 Free PMC article. Review.
Cited by
-
Extracellular Vesicles as Innovative Tool for Diagnosis, Regeneration and Protection against Neurological Damage.Int J Mol Sci. 2020 Sep 18;21(18):6859. doi: 10.3390/ijms21186859. Int J Mol Sci. 2020. PMID: 32962107 Free PMC article. Review.
-
Rethinking the bioavailability and cellular transport properties of S-adenosylmethionine.Cell Stress. 2021 Dec 6;6(1):1-5. doi: 10.15698/cst2022.01.261. eCollection 2022 Jan. Cell Stress. 2021. PMID: 35083422 Free PMC article.
-
Pharmacogenomic and Pharmacomicrobiomic Aspects of Drugs of Abuse.Genes (Basel). 2025 Mar 30;16(4):403. doi: 10.3390/genes16040403. Genes (Basel). 2025. PMID: 40282363 Free PMC article. Review.
-
Gut Microbiota as Important Mediator Between Diet and DNA Methylation and Histone Modifications in the Host.Nutrients. 2020 Feb 25;12(3):597. doi: 10.3390/nu12030597. Nutrients. 2020. PMID: 32106534 Free PMC article. Review.
-
Keto microbiota: A powerful contributor to host disease recovery.Rev Endocr Metab Disord. 2019 Dec;20(4):415-425. doi: 10.1007/s11154-019-09518-8. Rev Endocr Metab Disord. 2019. PMID: 31720986 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

