Expression Differences of Pigment Structural Genes and Transcription Factors Explain Flesh Coloration in Three Contrasting Kiwifruit Cultivars
- PMID: 28919902
- PMCID: PMC5586210
- DOI: 10.3389/fpls.2017.01507
Expression Differences of Pigment Structural Genes and Transcription Factors Explain Flesh Coloration in Three Contrasting Kiwifruit Cultivars
Abstract
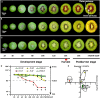
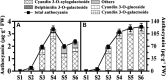
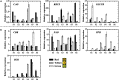
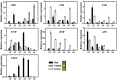
Fruits of kiwifruit cultivars (Actinidia chinensis and A. deliciosa) generally have green or yellow flesh when ripe. A small number of genotypes have red flesh but this coloration is usually restricted to the inner pericarp. Three kiwifruit cultivars having red ('Hongyang'), or yellow ('Jinnong-2'), or green ('Hayward') flesh were investigated for their color characteristics and pigment contents during development and ripening. The results show the yellow of the 'Jinnong-2' fruit is due to the combined effects of chlorophyll degradation and of beta-carotene accumulation. The red inner pericarps of 'Hongyang' fruit are due to anthocyanin accumulation. Expression differences of the pathway genes in the inner pericarps of the three different kiwifruits suggest that stay-green (SGR) controls the degradation of chlorophylls, while lycopene beta-cyclase (LCY-β) controls the biosynthesis of beta-carotene. The abundance of anthocyanin in the inner pericarps of the 'Hongyang' fruit is the results of high expressions of UDP flavonoid glycosyltransferases (UFGT). At the same time, expressions of anthocyanin transcription factors show that AcMYBF110 expression parallels changes in anthocyanin concentration, so seems to be a key R2R3 MYB, regulating anthocyanin biosynthesis. Further, transient color assays reveal that AcMYBF110 can autonomously induce anthocyanin accumulation in Nicotiana tabacum leaves by activating the transcription of dihydroflavonol 4-reductase (NtDFR), anthocyanidin synthase (NtANS) and NtUFGT. For basic helix-loop-helix proteins (bHLHs) and WD-repeat proteins (WD40s), expression differences show these may depend on AcMYBF110 forming a MYB-bHLH-WD40 complex to regulate anthocyanin biosynthesis, instead of it having a direct involvement.
Keywords: color diversity; expression differences; kiwifruit; structural genes; transcription factors.
Figures




References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

