Hepatic S6K1 Partially Regulates Lifespan of Mice with Mitochondrial Complex I Deficiency
- PMID: 28919908
- PMCID: PMC5585733
- DOI: 10.3389/fgene.2017.00113
Hepatic S6K1 Partially Regulates Lifespan of Mice with Mitochondrial Complex I Deficiency
Erratum in
-
Corrigendum: Hepatic S6K1 Partially Regulates Lifespan of Mice with Mitochondrial Complex I Deficiency.Front Genet. 2017 Dec 22;8:221. doi: 10.3389/fgene.2017.00221. eCollection 2017. Front Genet. 2017. PMID: 29285026 Free PMC article.
Abstract
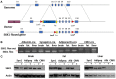
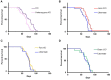
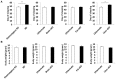
The inactivation of ribosomal protein S6 kinase 1 (S6K1) recapitulates aspects of caloric restriction and mTORC1 inhibition to achieve prolonged longevity in invertebrate and mouse models. In addition to delaying normative aging, inhibition of mTORC1 extends the shortened lifespan of yeast, fly, and mouse models with severe mitochondrial disease. Here we tested whether disruption of S6K1 can recapitulate the beneficial effects of mTORC1 inhibition in the Ndufs4 knockout (NKO) mouse model of Leigh Syndrome caused by Complex I deficiency. These NKO mice develop profound neurodegeneration resulting in brain lesions and death around 50-60 days of age. Our results show that liver-specific, as well as whole body, S6K1 deletion modestly prolongs survival and delays onset of neurological symptoms in NKO mice. In contrast, we observed no survival benefit in NKO mice specifically disrupted for S6K1 in neurons or adipocytes. Body weight was reduced in WT mice upon disruption of S6K1 in adipocytes or whole body, but not altered when S6K1 was disrupted only in neurons or liver. Taken together, these data indicate that decreased S6K1 activity in liver is sufficient to delay the neurological and survival defects caused by deficiency of Complex I and suggest that mTOR signaling can modulate mitochondrial disease and metabolism via cell non-autonomous mechanisms.
Keywords: S6K1; lifespan; liver; mTORC1; mitochondrial disease.
Figures
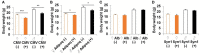
Similar articles
-
EPRS is a critical mTORC1-S6K1 effector that influences adiposity in mice.Nature. 2017 Feb 16;542(7641):357-361. doi: 10.1038/nature21380. Epub 2017 Feb 8. Nature. 2017. PMID: 28178239 Free PMC article.
-
Acarbose suppresses symptoms of mitochondrial disease in a mouse model of Leigh syndrome.Nat Metab. 2023 Jun;5(6):955-967. doi: 10.1038/s42255-023-00815-w. Epub 2023 Jun 26. Nat Metab. 2023. PMID: 37365290
-
Inhibition of mitochondrial complex 1 by the S6K1 inhibitor PF-4708671 partly contributes to its glucose metabolic effects in muscle and liver cells.J Biol Chem. 2019 Aug 9;294(32):12250-12260. doi: 10.1074/jbc.RA119.008488. Epub 2019 Jun 26. J Biol Chem. 2019. PMID: 31243102 Free PMC article.
-
Angiotensin II blockade: how its molecular targets may signal to mitochondria and slow aging. Coincidences with calorie restriction and mTOR inhibition.Am J Physiol Heart Circ Physiol. 2015 Jul 1;309(1):H15-44. doi: 10.1152/ajpheart.00459.2014. Epub 2015 May 1. Am J Physiol Heart Circ Physiol. 2015. PMID: 25934099 Review.
-
Growth signaling and longevity in mouse models.BMB Rep. 2019 Jan;52(1):70-85. doi: 10.5483/BMBRep.2019.52.1.299. BMB Rep. 2019. PMID: 30545442 Free PMC article. Review.
Cited by
-
Ribosomal S6 kinase 1 regulates inflammaging via the senescence secretome.Nat Aging. 2024 Nov;4(11):1544-1561. doi: 10.1038/s43587-024-00695-z. Epub 2024 Aug 29. Nat Aging. 2024. PMID: 39210150 Free PMC article.
-
Iron status influences mitochondrial disease progression in Complex I-deficient mice.Elife. 2023 Feb 17;12:e75825. doi: 10.7554/eLife.75825. Elife. 2023. PMID: 36799301 Free PMC article.
-
Cross-comparison of systemic and tissue-specific metabolomes in a mouse model of Leigh syndrome.Metabolomics. 2021 Nov 18;17(12):101. doi: 10.1007/s11306-021-01854-8. Metabolomics. 2021. PMID: 34792662
-
A novel role for GalNAc-T2 dependent glycosylation in energy homeostasis.Mol Metab. 2022 Jun;60:101472. doi: 10.1016/j.molmet.2022.101472. Epub 2022 Mar 15. Mol Metab. 2022. PMID: 35304331 Free PMC article.
-
An energetics perspective on geroscience: mitochondrial protonmotive force and aging.Geroscience. 2021 Aug;43(4):1591-1604. doi: 10.1007/s11357-021-00365-7. Epub 2021 Apr 17. Geroscience. 2021. PMID: 33864592 Free PMC article.
References
-
- Caccamo A., De Pinto V., Messina A., Branca C., Oddo S. (2014). Genetic reduction of mammalian target of rapamycin ameliorates Alzheimer's disease-like cognitive and pathological deficits by restoring hippocampal gene expression signature. J. Neurosci. 34, 7988–7998. 10.1523/JNEUROSCI.0777-14.2014 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

