Immunocytological analysis of meiotic recombination in two anole lizards (Squamata, Dactyloidae)
- PMID: 28919954
- PMCID: PMC5599703
- DOI: 10.3897/CompCytogen.v11i1.10916
Immunocytological analysis of meiotic recombination in two anole lizards (Squamata, Dactyloidae)
Abstract
Although the evolutionary importance of meiotic recombination is not disputed, the significance of interspecies differences in the recombination rates and recombination landscapes remains under-appreciated. Recombination rates and distribution of chiasmata have been examined cytologically in many mammalian species, whereas data on other vertebrates are scarce. Immunolocalization of the protein of the synaptonemal complex (SYCP3), centromere proteins and the mismatch-repair protein MLH1 was used, which is associated with the most common type of recombination nodules, to analyze the pattern of meiotic recombination in the male of two species of iguanian lizards, Anolis carolinensis Voigt, 1832 and Deiroptyx coelestinus (Cope, 1862). These species are separated by a relatively long evolutionary history although they retain the ancestral iguanian karyotype. In both species similar and extremely uneven distributions of MLH1 foci along the macrochromosome bivalents were detected: approximately 90% of crossovers were located at the distal 20% of the chromosome arm length. Almost total suppression of recombination in the intermediate and proximal regions of the chromosome arms contradicts the hypothesis that "homogenous recombination" is responsible for the low variation in GC content across the anole genome. It also leads to strong linkage disequilibrium between the genes located in these regions, which may benefit conservation of co-adaptive gene arrays responsible for the ecological adaptations of the anoles.
Keywords: Anolis; Deiroptyx; Reptilia; Synaptonemal complex; chromosomes; crossing over; lizard.
Figures
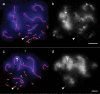
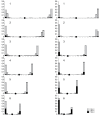
Similar articles
-
Heteromorphism of "Homomorphic" Sex Chromosomes in Two Anole Species (Squamata, Dactyloidae) Revealed by Synaptonemal Complex Analysis.Cytogenet Genome Res. 2017;151(2):89-95. doi: 10.1159/000460829. Epub 2017 Mar 18. Cytogenet Genome Res. 2017. PMID: 28315859
-
Male Meiotic Recombination in the Steppe Agama, Trapelus sanguinolentus (Agamidae, Iguania, Reptilia).Cytogenet Genome Res. 2019;157(1-2):107-114. doi: 10.1159/000496078. Epub 2019 Jan 25. Cytogenet Genome Res. 2019. PMID: 30677759
-
New insights into sex chromosome evolution in anole lizards (Reptilia, Dactyloidae).Chromosoma. 2017 Mar;126(2):245-260. doi: 10.1007/s00412-016-0585-6. Epub 2016 Mar 22. Chromosoma. 2017. PMID: 27001473
-
Ultrastructural analysis of meiotic recombination and chiasma formation.Tokai J Exp Clin Med. 1986 Dec;11(6):415-36. Tokai J Exp Clin Med. 1986. PMID: 3328325 Review.
-
Cytological analysis of interference in mouse meiosis.Methods Mol Biol. 2009;558:355-82. doi: 10.1007/978-1-60761-103-5_21. Methods Mol Biol. 2009. PMID: 19685335 Review.
Cited by
-
Meiotic chromosome dynamics and double strand break formation in reptiles.Front Cell Dev Biol. 2022 Oct 12;10:1009776. doi: 10.3389/fcell.2022.1009776. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36313577 Free PMC article.
-
Whole-chromosome fusions in the karyotype evolution of Sceloporus (Iguania, Reptilia) are more frequent in sex chromosomes than autosomes.Philos Trans R Soc Lond B Biol Sci. 2021 Sep 13;376(1833):20200099. doi: 10.1098/rstb.2020.0099. Epub 2021 Jul 26. Philos Trans R Soc Lond B Biol Sci. 2021. PMID: 34304596 Free PMC article.
-
Genomes of two Extinct-in-the-Wild reptiles from Christmas Island reveal distinct evolutionary histories and conservation insights.Mol Ecol Resour. 2025 Jul;25(5):e13780. doi: 10.1111/1755-0998.13780. Epub 2023 Mar 21. Mol Ecol Resour. 2025. PMID: 36872490 Free PMC article.
-
Cytological Analysis of Crossover Frequency and Distribution in Male Meiosis of Cardueline Finches (Fringillidae, Aves).Animals (Basel). 2023 Nov 23;13(23):3624. doi: 10.3390/ani13233624. Animals (Basel). 2023. PMID: 38066976 Free PMC article.
-
Sex-chromosome evolution in frogs: what role for sex-antagonistic genes?Philos Trans R Soc Lond B Biol Sci. 2021 Aug 30;376(1832):20200094. doi: 10.1098/rstb.2020.0094. Epub 2021 Jul 12. Philos Trans R Soc Lond B Biol Sci. 2021. PMID: 34247502 Free PMC article.
References
-
- Alföldi J, Di Palma F, Grabherr M, Williams C, Kong L, Mauceli E, et al. (2011) The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature 477(7366): 587–591. https://doi.org/10.1038/nature10390 - DOI - PMC - PubMed
-
- Barton NH, Otto SP. (2005) Evolution of recombination due to random drift. Genetics 169(4): 2353–2370. https://doi.org/10.1534/genetics.104.032821 - DOI - PMC - PubMed
-
- Basheva EA, Bidau CJ, Borodin PM. (2008) General pattern of meiotic recombination in male dogs estimated by MLH1 and RAD51 immunolocalization. Chromosome Research 16(5): 709–719. https://doi.org/10.1007/s10577-008-1221-y - DOI - PubMed
-
- Beçak W, Beçak ML, Nazareth HRS, Ohno S. (1964) Close karyological kinship between the reptilian suborder Serpentes and the class Aves. Chromosoma 15(5): 606–617. https://doi.org/10.1007/BF00319994 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
