Identification of a HLA-A*0201-restricted immunogenic epitope from the universal tumor antigen DEPDC1
- PMID: 28919988
- PMCID: PMC5593712
- DOI: 10.1080/2162402X.2017.1313371
Identification of a HLA-A*0201-restricted immunogenic epitope from the universal tumor antigen DEPDC1
Erratum in
-
Corrigendum.Oncoimmunology. 2017 Dec 4;7(1):e1412885. doi: 10.1080/2162402X.2017.1412885. eCollection 2017. Oncoimmunology. 2017. PMID: 29296545 Free PMC article.
Abstract
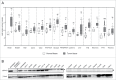
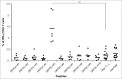
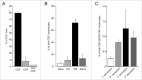
The identification of universal tumor-specific antigens shared between multiple patients and/or multiple tumors is of great importance to overcome the practical limitations of personalized cancer immunotherapy. Recent studies support the involvement of DEPDC1 in many aspects of cancer traits, such as cell proliferation, resistance to induction of apoptosis and cell invasion, suggesting that it may play key roles in the oncogenic process. In this study, we report that DEPDC1 expression is upregulated in most types of human tumors, and closely linked to a poorer prognosis; therefore, it might be regarded as a novel universal oncoantigen potentially suitable for targeting many different cancers. In this regard, we report the identification of a HLA-A*0201 allele-restricted immunogenic DEPDC1-derived epitope, which is able to induce cytotoxic T lymphocytes (CTL) exerting a strong and specific functional response in vitro toward not only peptide-loaded cells but also triple negative breast cancer (TNBC) cells endogenously expressing the DEPDC1 protein. Such CTL are also therapeutically active against human TNBC xenografts in vivo upon adoptive transfer in immunodeficient mice. Overall, these data provide evidence that this DEPDC1-derived antigenic epitope can be exploited as a new tool for developing immunotherapeutic strategies for HLA-A*0201 patients with TNBC, and potentially many other cancers.
Keywords: Cancer immunotherapy; DEPDC1; cytotoxic T lymphocytes; triple negative breast cancer; tumor antigen.
Figures



Similar articles
-
Bladder cancer-associated cancer-testis antigen-derived long peptides encompassing both CTL and promiscuous HLA class II-restricted Th cell epitopes induced CD4+ T cells expressing converged T-cell receptor genes in vitro.Oncoimmunology. 2018 Jan 5;7(4):e1415687. doi: 10.1080/2162402X.2017.1415687. eCollection 2018. Oncoimmunology. 2018. PMID: 29632734 Free PMC article.
-
Identification of HLA-A24-restricted CTL epitope from cancer-testis antigen, NY-ESO-1, and induction of a specific antitumor immune response.Clin Cancer Res. 2004 Feb 1;10(3):890-6. doi: 10.1158/1078-0432.ccr-1086-3. Clin Cancer Res. 2004. PMID: 14871964
-
DEPDC1, negatively regulated by miR-26b, facilitates cell proliferation via the up-regulation of FOXM1 expression in TNBC.Cancer Lett. 2019 Feb 1;442:242-251. doi: 10.1016/j.canlet.2018.11.003. Epub 2018 Nov 9. Cancer Lett. 2019. PMID: 30419349
-
Immunodominance across HLA polymorphism: implications for cancer immunotherapy.J Immunother. 1998 Jan;21(1):1-16. J Immunother. 1998. PMID: 9456431 Review.
-
Cancer immunotherapy using novel tumor-associated antigenic peptides identified by genome-wide cDNA microarray analyses.Cancer Sci. 2015 May;106(5):505-11. doi: 10.1111/cas.12650. Epub 2015 Apr 1. Cancer Sci. 2015. PMID: 25726868 Free PMC article. Review.
Cited by
-
Glycolysis-related gene expression profiling serves as a novel prognosis risk predictor for human hepatocellular carcinoma.Sci Rep. 2021 Sep 23;11(1):18875. doi: 10.1038/s41598-021-98381-2. Sci Rep. 2021. PMID: 34556750 Free PMC article.
-
Adoptive cell therapy of triple negative breast cancer with redirected cytokine-induced killer cells.Oncoimmunology. 2020 Jun 11;9(1):1777046. doi: 10.1080/2162402X.2020.1777046. Oncoimmunology. 2020. PMID: 32923140 Free PMC article.
-
Targeting DEP domain containing 1 in anaplastic thyroid carcinoma: Implications for stemness regulation and malignant phenotype suppression.Heliyon. 2024 Feb 29;10(5):e27150. doi: 10.1016/j.heliyon.2024.e27150. eCollection 2024 Mar 15. Heliyon. 2024. PMID: 38449652 Free PMC article.
-
Roles of DEPDC1 in various types of cancer (Review).Oncol Lett. 2024 Aug 29;28(5):518. doi: 10.3892/ol.2024.14651. eCollection 2024 Nov. Oncol Lett. 2024. PMID: 39296974 Free PMC article. Review.
-
Cancer-Testis Antigen Peptide Vaccine for Cancer Immunotherapy: Progress and Prospects.Transl Oncol. 2019 May;12(5):733-738. doi: 10.1016/j.tranon.2019.02.008. Epub 2019 Mar 14. Transl Oncol. 2019. PMID: 30877975 Free PMC article. Review.
References
-
- Neves H, Kwok HF. Recent advances in the field of anti-cancer immunotherapy. BBA Clin 2015; 3:280-8; PMID:26673349; https://doi.org/10.1016/j.bbacli.2015.04.001 - DOI - PMC - PubMed
-
- Mahoney KM, Rennert PD, Freeman GJ. Combination cancer immunotherapy and new immunomodulatory targets. Nat Rev Drug Discov 2015; 14(8):561-84; PMID:26228759; https://doi.org/10.1038/nrd4591 - DOI - PubMed
-
- Taube JM, Klein A, Brahmer JR, Xu H, Pan X, Kim JH, Chen L, Pardoll DM, Topalian SL, Anders RA et al.. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin Cancer Res. 2014; 20(19):5064-74; PMID:24714771; https://doi.org/10.1158/1078-0432.CCR-13-3271 - DOI - PMC - PubMed
-
- Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS et al.. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015; 348(6230):124-8; PMID:25765070; https://doi.org/10.1126/science.aaa1348 - DOI - PMC - PubMed
-
- McGranahan N, Furness AJS, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, Jamal-Hanjani M, Wilson GA, Birkbak NJ, Hiley CT et al.. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016; 351(6280):1463-9; PMID:26940869; https://doi.org/10.1126/science.aaf1490 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
