Neuroblastoma cells undergo transcriptomic alterations upon dissemination into the bone marrow and subsequent tumor progression
- PMID: 28921546
- PMCID: PMC5725737
- DOI: 10.1002/ijc.31053
Neuroblastoma cells undergo transcriptomic alterations upon dissemination into the bone marrow and subsequent tumor progression
Abstract
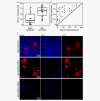
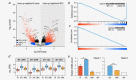
Neuroblastoma is the most common extracranial solid tumor in childhood. The vast majority of metastatic (M) stage patients present with disseminated tumor cells (DTCs) in the bone marrow (BM) at diagnosis and relapse. Although these cells represent a major obstacle in the treatment of neuroblastoma patients, insights into their expression profile remained elusive. The present RNA-Seq study of stage 4/M primary tumors, enriched BM-derived diagnostic and relapse DTCs, as well as the corresponding BM-derived mononuclear cells (MNCs) from 53 patients revealed 322 differentially expressed genes in DTCs as compared to the tumors (q < 0.001, |log2 FC|>2). Particularly, the levels of transcripts encoded by mitochondrial DNA were elevated in DTCs, whereas, for example, genes involved in angiogenesis were downregulated. Furthermore, 224 genes were highly expressed in DTCs and only slightly, if at all, in MNCs (q < 8 × 10-75 log2 FC > 6). Interestingly, we found the transcriptome of relapse DTCs largely resembling those of diagnostic DTCs with only 113 differentially expressed genes under relaxed cut-offs (q < 0.01, |log2 FC|>0.5). Notably, relapse DTCs showed a positional enrichment of 31 downregulated genes on chromosome 19, including five tumor suppressor genes: SIRT6, BBC3/PUMA, STK11, CADM4 and GLTSCR2. This first RNA-Seq analysis of neuroblastoma DTCs revealed their unique expression profile in comparison to the tumors and MNCs, and less pronounced differences between diagnostic and relapse DTCs. The latter preferentially affected downregulation of genes encoded by chromosome 19. As these alterations might be associated with treatment failure and disease relapse, further functional studies on DTCs should be considered.
Keywords: RNA-Seq; bone marrow; disseminated tumor cells; neuroblastoma.
© 2017 The Authors International Journal of Cancer published by John Wiley & Sons Ltd on behalf of UICC.
Figures




References
-
- Matthay KK, Maris JM, Schleiermacher G, et al. Neuroblastoma. Nat Rev Dis Primers 2016;2:16078 - PubMed
-
- Ambros IM, Zellner A, Roald B, et al. Role of ploidy, chromosome 1p, and Schwann cells in the maturation of neuroblastoma. N Engl J Med 1996;334:1505–11. - PubMed
-
- Molenaar JJ, Koster J, Zwijnenburg DA, et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature 2012;483:589–93. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

