Estimate your dose: RADDOSE-3D
- PMID: 28921782
- PMCID: PMC5734275
- DOI: 10.1002/pro.3302
Estimate your dose: RADDOSE-3D
Abstract
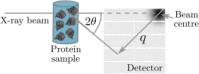
We present the current status of RADDOSE-3D, a software tool allowing the estimation of the dose absorbed in a macromolecular crystallography diffraction experiment. The code allows a temporal and spatial dose contour map to be calculated for a crystal of any geometry and size as it is rotated in an X-ray beam, and gives several summary dose values: among them diffraction weighted dose. This allows experimenters to plan data collections which will minimize radiation damage effects by spreading the absorbed dose more homogeneously, and thus to optimize the use of their crystals. It also allows quantitative comparisons between different radiation damage studies, giving a universal "x-axis" against which to plot various metrics.
Keywords: absorption coefficients; beam profile; diffraction weighted dose; dose; radiation damage.
© 2017 The Protein Society.
Figures







References
-
- Blake CCF, Phillips DC (1962) Effects of X‐irradiaton on single crystals of myoglobin. Proceedings of the Symposium on the Biological Effects of Ionising radiation at the Molecular Level. International Atomic Energy Agency, Vienna, pp. 183–191.
-
- Teng T, Moffat K (2000) Primary radiation damage of protein crystals by intense synchrotron radiation. J Synchr Radiat 7:313–317. - PubMed
-
- Kmetko J, Husseini NS, Naides M, Kalinin Y, Thorne RE (2006) Quantifying X‐ray radiation damage in protein crystals at cryogenic temperatures. Acta Cryst D62:1030–1038. - PubMed
-
- Diederichs K (2006) Some aspects of quantitative analysis and correction of radiation damage. Acta Cryst D62:96–101. - PubMed
-
- De la Mora E, Carmichael I, Garman EF (2011) Effective scavenging at cryotemperatures: Further increasing the dose tolerance of protein crystals. J Synchr Radiat 18:346–357. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

