Rice Phytochrome-Interacting Factor-Like1 (OsPIL1) is involved in the promotion of chlorophyll biosynthesis through feed-forward regulatory loops
- PMID: 28922754
- PMCID: PMC5853433
- DOI: 10.1093/jxb/erx231
Rice Phytochrome-Interacting Factor-Like1 (OsPIL1) is involved in the promotion of chlorophyll biosynthesis through feed-forward regulatory loops
Abstract
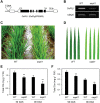
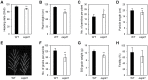
In phototrophic plants, the highly conserved and tightly regulated process of chlorophyll (Chl) biosynthesis comprises multi-step reactions involving more than 15 enzymes. Since the efficiency of Chl biosynthesis strongly affects plant productivity, understanding the underlying regulatory mechanisms in crop plants can be useful for strategies to increase grain and biomass yields. Here, we show that rice (Oryza sativa) Phytochrome-Interacting Factor-Like1 (OsPIL1), a basic helix-loop-helix transcription factor, promotes Chl biosynthesis. The T-DNA insertion knockdown ospil1 mutant showed a pale-green phenotype when grown in a natural paddy field. Transcriptome analysis revealed that several genes responsible for Chl biosynthesis and photosynthesis were significantly down-regulated in ospil1 leaves. Using promoter binding and transactivation assays, we found that OsPIL1 binds to the promoters of two Chl biosynthetic genes, OsPORB and OsCAO1, and promotes their transcription. In addition, OsPIL1 directly up-regulates the expression of two transcription factor genes, GOLDEN2-LIKE1 (OsGLK1) and OsGLK2. OsGLK1 and OsGLK2 both bind to the promoters of OsPORB and OsCAO1, as well as some of genes encoding the light-harvesting complex of photosystems, probably promoting their transcription. Thus, OsPIL1 is involved in the promotion of Chl biosynthesis by up-regulating the transcription of OsPORB and OsCAO1 via trifurcate feed-forward regulatory loops involving two OsGLKs.
Keywords: Chlorophyll biosynthesis; OsCAO1; OsGLK; OsPIL1; OsPORB; rice; transcriptional regulation.
© The Author 2017. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures



References
-
- Al-Sady B, Ni W, Kircher S, Schäfer E, Quail PH. 2006. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Molecular Cell 23, 439–446. - PubMed
-
- Blankenship RE. 1992. Origin and early evolution of photosynthesis. Photosynthesis Research 33, 91–111. - PubMed
-
- Castillon A, Shen H, Huq E. 2007. Phytochrome interacting factors: central players in phytochrome-mediated light signaling networks. Trends in Plant Science 12, 514–521. - PubMed
-
- Chen H, Cheng Z, Ma X et al. . 2013. A knockdown mutation of YELLOW-GREEN LEAF2 blocks chlorophyll biosynthesis in rice. Plant Cell Reports 32, 1855–1867. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

