A pentatricopeptide repeat protein restores nap cytoplasmic male sterility in Brassica napus
- PMID: 28922764
- PMCID: PMC5853434
- DOI: 10.1093/jxb/erx239
A pentatricopeptide repeat protein restores nap cytoplasmic male sterility in Brassica napus
Abstract
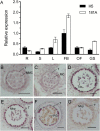
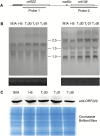
Two forms of male-sterile cytoplasm, designated nap and pol, are found in the oilseed rape species, Brassica napus. The nap cytoplasm is observed in most B. napus varieties, and it confers male sterility on a limited number of cultivars that lack the corresponding restorer gene, Rfn. In the present study, using linkage analysis in combination with 5652 BC1 progeny derived from a cross between a nap cytoplasmic male sterility (CMS) line 181A and a restorer line H5, we delimited the Rfn gene to a 10.5 kb region on chromosome A09, which contained three putative ORFs. Complementation by transformation rescue revealed that the introduction of ORF2, which encodes a pentatricopeptide repeat (PPR) protein, resulted in the recovery of fertility of nap CMS plants. Expression analysis suggested that the Rfn was highly expressed in flower buds and it was preferentially expressed in the tapetum and meiocytes during anther development. Further RNA gel blots and immunodetection suggested that the Rfn gene may play a complicated role in restoring the nap CMS. Our work laid the foundation for dissecting the molecular basis of CMS fertility restoration and the nuclear-mitochondrial interactions in CMS/Rf systems.
Keywords: Brassica napus; cytoplasmic male sterility; map-based cloning; mitochondria; pentatricopeptide repeat protein; restorer of fertility (Rf).
© The Author 2017. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures



Similar articles
-
Comparative genomic analysis of the compound Brassica napus Rf locus.BMC Genomics. 2016 Oct 26;17(1):834. doi: 10.1186/s12864-016-3117-0. BMC Genomics. 2016. PMID: 27782804 Free PMC article.
-
A mitochondria-localized pentatricopeptide repeat protein is required to restore hau cytoplasmic male sterility in Brassica napus.Theor Appl Genet. 2021 May;134(5):1377-1386. doi: 10.1007/s00122-021-03777-3. Epub 2021 Mar 16. Theor Appl Genet. 2021. PMID: 33725137
-
Genetic characterization of a pentatricopeptide repeat protein gene, orf687, that restores fertility in the cytoplasmic male-sterile Kosena radish.Plant J. 2003 May;34(4):407-15. doi: 10.1046/j.1365-313x.2003.01735.x. Plant J. 2003. PMID: 12753581
-
An overview of cytoplasmic male sterility in Brassica napus.Funct Plant Biol. 2025 May;52:FP24337. doi: 10.1071/FP24337. Funct Plant Biol. 2025. PMID: 40310995 Review.
-
Mechanism and Utilization of Ogura Cytoplasmic Male Sterility in Cruciferae Crops.Int J Mol Sci. 2022 Aug 13;23(16):9099. doi: 10.3390/ijms23169099. Int J Mol Sci. 2022. PMID: 36012365 Free PMC article. Review.
Cited by
-
Isolation and identification of a vegetative organ-specific promoter from maize.Physiol Mol Biol Plants. 2019 Jan;25(1):277-287. doi: 10.1007/s12298-018-0546-z. Epub 2018 Jun 4. Physiol Mol Biol Plants. 2019. PMID: 30804649 Free PMC article.
-
Development and Validation of Markers for the Fertility Restorer Gene Rf1 in Sunflower.Int J Mol Sci. 2019 Mar 13;20(6):1260. doi: 10.3390/ijms20061260. Int J Mol Sci. 2019. PMID: 30871146 Free PMC article.
-
Assembly and analysis of the mitochondrial genome of Prunella vulgaris.Front Plant Sci. 2023 Aug 2;14:1237822. doi: 10.3389/fpls.2023.1237822. eCollection 2023. Front Plant Sci. 2023. PMID: 37600185 Free PMC article.
-
Transcript levels of orf288 are associated with the hau cytoplasmic male sterility system and altered nuclear gene expression in Brassica juncea.J Exp Bot. 2018 Jan 23;69(3):455-466. doi: 10.1093/jxb/erx443. J Exp Bot. 2018. PMID: 29301015 Free PMC article.
-
Molecular Analysis Uncovers the Mechanism of Fertility Restoration in Temperature-Sensitive Polima Cytoplasmic Male-Sterile Brassica napus.Int J Mol Sci. 2021 Nov 18;22(22):12450. doi: 10.3390/ijms222212450. Int J Mol Sci. 2021. PMID: 34830333 Free PMC article.
References
-
- Arnal N, Quadrado M, Simon M, Mireau H. 2014. A restorer-of-fertility like pentatricopeptide repeat gene directs ribonucleolytic processing within the coding sequence of rps3-rpl16 and orf240a mitochondrial transcripts in Arabidipsis thaliana. The Plant Journal 78, 134–145. - PubMed
-
- Barkan A, Small I. 2014. Pentatricopeptide repeat proteins in plants. Annual Review of Plant Biology 65, 415–442. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

