Divergence of the Venom Exogene Repertoire in Two Sister Species of Turriconus
- PMID: 28922871
- PMCID: PMC5604253
- DOI: 10.1093/gbe/evx157
Divergence of the Venom Exogene Repertoire in Two Sister Species of Turriconus
Abstract
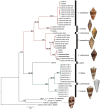
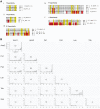
The genus Conus comprises approximately 700 species of venomous marine cone snails that are highly efficient predators of worms, snails, and fish. In evolutionary terms, cone snails are relatively young with the earliest fossil records occurring in the Lower Eocene, 55 Ma. The rapid radiation of cone snail species has been accompanied by remarkably high rates of toxin diversification. To shed light on the molecular mechanisms that accompany speciation, we investigated the toxin repertoire of two sister species, Conus andremenezi and Conus praecellens, that were until recently considered a single variable species. A total of 196 and 250 toxin sequences were identified in the venom gland transcriptomes of C. andremenezi and C. praecellens belonging to 25 and 29 putative toxin gene superfamilies, respectively. Comparative analysis with closely (Conus tribblei and Conus lenavati) and more distantly related species (Conus geographus) suggests that speciation is associated with significant diversification of individual toxin genes (exogenes) whereas the expression pattern of toxin gene superfamilies within lineages remains largely conserved. Thus, changes within individual toxin sequences can serve as a sensitive indicator for recent speciation whereas changes in the expression pattern of gene superfamilies are likely to reflect more dramatic differences in a species' interaction with its prey, predators, and competitors.
Keywords: conotoxins; exogenes; speciation; venom evolution.
© The Author 2017. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

