Ancient Occasional Host Switching of Maternally Transmitted Bacterial Symbionts of Chemosynthetic Vesicomyid Clams
- PMID: 28922872
- PMCID: PMC5604134
- DOI: 10.1093/gbe/evx166
Ancient Occasional Host Switching of Maternally Transmitted Bacterial Symbionts of Chemosynthetic Vesicomyid Clams
Abstract
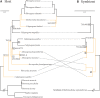
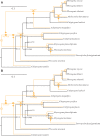
Vesicomyid clams in deep-sea chemosynthetic ecosystems harbor sulfur-oxidizing bacteria in their gill epithelial cells. These symbionts, which are vertically transmitted, are species-specific and thought to have cospeciated with their hosts. However, recent studies indicate incongruent phylogenies between some vesicomyid clams and their symbionts, suggesting that symbionts are horizontally transmitted. To more precisely understand the evolution of vesicomyid clams and their symbionts, we compared the evolution of vesicomyid clams and their symbionts through phylogenetic analyses using multi-gene data sets. Many clades in the phylogenetic trees of 13 host species (Abyssogena mariana, Ab. phaseoliformis, Akebiconcha kawamurai, Calyptogena fausta, C. laubieri, C. magnifica, C. nautilei, C. pacifica, Isorropodon fossajaponicum, Phreagena kilmeri, Ph. okutanii, Ph. soyoae, and Pliocardia stearnsii) and their symbionts were well resolved. Six of the 13 host-symbiont pairs (C. fausta, C. magnifica, C. pacifica, Ph. kilmeri, Ph. okutanii, and Ph. soyoae, and their respective symbionts) showed topological congruence. However, the remaining seven pairs (Ak. kawamurai, Ab mariana, Ab. phaseoliformis, C. laubieri, C. nautilei, I. fossajaponicum, and Pl. stearnsii and their corresponding symbionts) showed incongruent topologies, which were supported by the approximately unbiased and Bayes factor tests. Coevolution analyses indicated that six pairs cospeciated, whereas host switching events occurred in the remaining seven pairs. Markedly, multiple host switching events may have occurred in the lineages from the common ancestral symbiont of C. pacifica and C. fausta. Our phylogenetic and coevolution analyses provide additional evidence for host switching during the evolution of vesicomyids.
Keywords: cospeciation; endosymbiosis; host switching; phylogenetic tree; vertical transmission.
© The Author 2017. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures
Similar articles
-
Loss of genes for DNA recombination and repair in the reductive genome evolution of thioautotrophic symbionts of Calyptogena clams.BMC Evol Biol. 2011 Oct 3;11:285. doi: 10.1186/1471-2148-11-285. BMC Evol Biol. 2011. PMID: 21966992 Free PMC article.
-
Host hybridization as a potential mechanism of lateral symbiont transfer in deep-sea vesicomyid clams.Mol Ecol. 2019 Nov;28(21):4697-4708. doi: 10.1111/mec.15224. Epub 2019 Sep 23. Mol Ecol. 2019. PMID: 31478269 Free PMC article.
-
Lateral symbiont acquisition in a maternally transmitted chemosynthetic clam endosymbiosis.Mol Biol Evol. 2008 Apr;25(4):673-87. doi: 10.1093/molbev/msn010. Epub 2008 Jan 12. Mol Biol Evol. 2008. PMID: 18192696
-
Bacterial endosymbioses in Solemya (Mollusca: Bivalvia)--model systems for studies of symbiont-host adaptation.Antonie Van Leeuwenhoek. 2006 Nov;90(4):343-60. doi: 10.1007/s10482-006-9086-6. Epub 2006 Oct 7. Antonie Van Leeuwenhoek. 2006. PMID: 17028934 Review.
-
Symbioses between deep-sea mussels (Mytilidae: Bathymodiolinae) and chemosynthetic bacteria: diversity, function and evolution.C R Biol. 2009 Feb-Mar;332(2-3):298-310. doi: 10.1016/j.crvi.2008.08.003. Epub 2008 Nov 25. C R Biol. 2009. PMID: 19281960 Review.
Cited by
-
Symbiont Community Composition in Rimicaris kairei Shrimps from Indian Ocean Vents with Notes on Mineralogy.Appl Environ Microbiol. 2022 Apr 26;88(8):e0018522. doi: 10.1128/aem.00185-22. Epub 2022 Apr 11. Appl Environ Microbiol. 2022. PMID: 35404070 Free PMC article.
-
Endosymbiont population genomics sheds light on transmission mode, partner specificity, and stability of the scaly-foot snail holobiont.ISME J. 2022 Sep;16(9):2132-2143. doi: 10.1038/s41396-022-01261-4. Epub 2022 Jun 17. ISME J. 2022. PMID: 35715703 Free PMC article.
-
Endosymbioses Have Shaped the Evolution of Biological Diversity and Complexity Time and Time Again.Genome Biol Evol. 2024 Jun 4;16(6):evae112. doi: 10.1093/gbe/evae112. Genome Biol Evol. 2024. PMID: 38813885 Free PMC article. Review.
-
Transmission strategies in a chemosynthetic symbiosis: detection and quantification of symbionts in host tissues and their environment.Proc Biol Sci. 2018 Oct 31;285(1890):20182157. doi: 10.1098/rspb.2018.2157. Proc Biol Sci. 2018. PMID: 30381385 Free PMC article.
-
Comparative mitochondrial genomic analyses of three chemosynthetic vesicomyid clams from deep-sea habitats.Ecol Evol. 2018 Jun 27;8(15):7261-7272. doi: 10.1002/ece3.4153. eCollection 2018 Aug. Ecol Evol. 2018. PMID: 30151147 Free PMC article.
References
-
- Barry JP, Whaling PJ, Kochevar RK.. 2007. Growth, production, and mortality of the chemosynthetic vesicomyid bivalve, Calyptogena kilmeri from cold seeps off central California. Mar Ecol. 28(1):169–182.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

