Comparative transcriptomics of female and male gametocytes in Plasmodium berghei and the evolution of sex in alveolates
- PMID: 28923023
- PMCID: PMC5604118
- DOI: 10.1186/s12864-017-4100-0
Comparative transcriptomics of female and male gametocytes in Plasmodium berghei and the evolution of sex in alveolates
Abstract
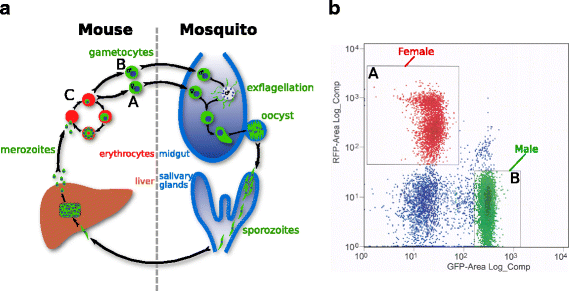
Background: The clinical symptoms of malaria are caused by the asexual replication of Plasmodium parasites in the blood of the vertebrate host. To spread to new hosts, however, the malaria parasite must differentiate into sexual forms, termed gametocytes, which are ingested by a mosquito vector. Sexual differentiation produces either female or male gametocytes, and involves significant morphological and biochemical changes. These transformations prepare gametocytes for the rapid progression to gamete formation and fertilisation, which occur within 20 min of ingestion. Here we present the transcriptomes of asexual, female, and male gametocytes in P. berghei, and a comprehensive statistically-based differential-expression analysis of the transcriptional changes that underpin this sexual differentiation.
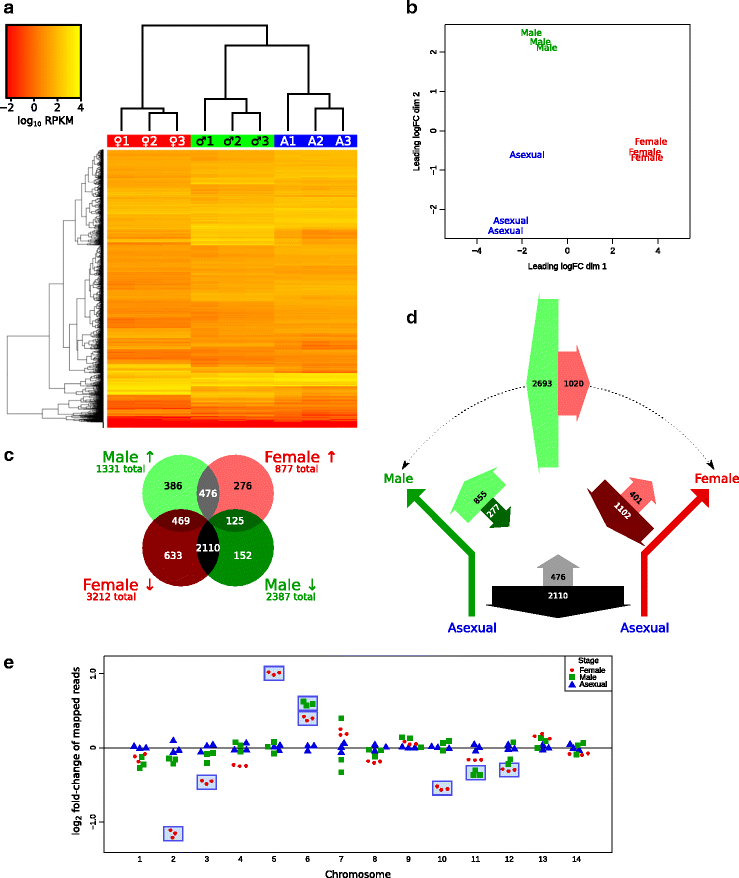
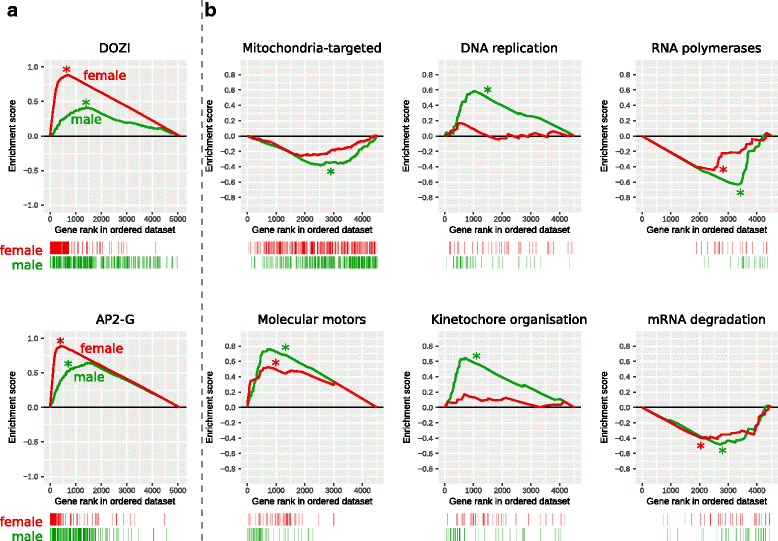
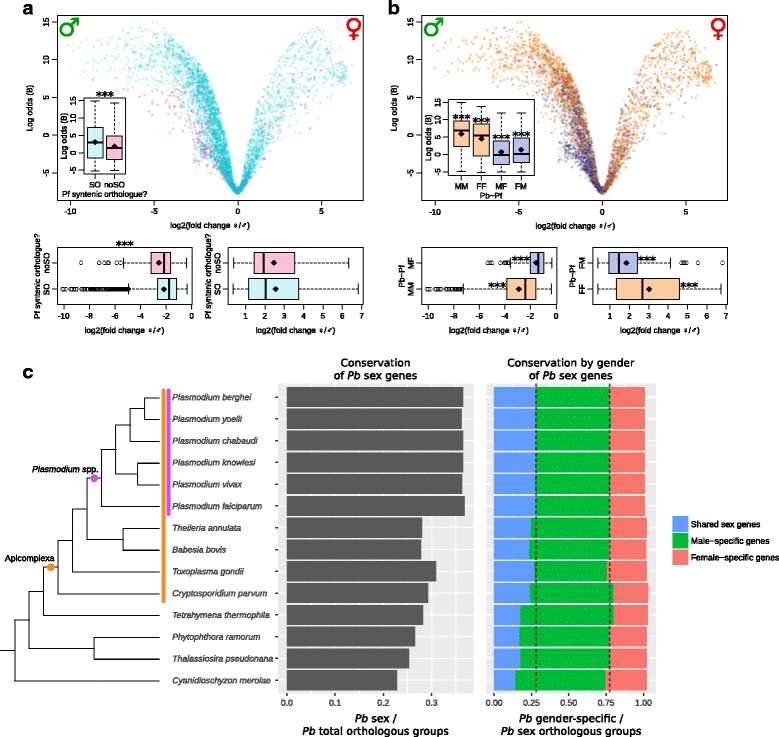
Results: RNA-seq analysis revealed numerous differences in the transcriptomes of female and male gametocytes compared to asexual stages. Overall, there is net downregulation of transcripts in gametocytes compared to asexual stages, with this trend more marked in female gametocytes. Our analysis identified transcriptional changes in previously-characterised gametocyte-specific pathways, which validated our approach. We also detected many previously-unreported female- and male-specific pathways and genes. Transcriptional biases in stage and gender were then used to investigate sex-specificity and sexual dimorphism of Plasmodium in an evolutionary context. Sex-related gene expression is well conserved between Plasmodium species, but relatively poorly conserved in related organisms outside this genus. This pattern of conservation is most evident in genes necessary for both male and female gametocyte formation. However, this trend is less pronounced for male-specific genes, which are more highly conserved outside the genus than genes specific to female development.
Conclusions: We characterised the transcriptional changes that are integral to the development of the female and male sexual forms of Plasmodium. These differential-expression patterns provide a vital insight into understanding the gender-specific characteristics of this essential stage that is the primary target for treatments that block parasite transmission. Our results also offer insight into the evolution of sex genes through Alveolata, and suggest that many Plasmodium sex genes evolved within the genus. We further hypothesise that male gametocytes co-opted pre-existing cellular machinery in their evolutionary history, whereas female gametocytes evolved more through the development of novel, parasite-specific pathways.
Keywords: Evolution; Gametocyte; Malaria; Next-generation sequencing; Plasmodium; Plasmodium berghei; RNA-seq; Sex; Transcriptome.
Conflict of interest statement
Ethics approval and consent to participate
Experiments were performed in conformity with the Australian Prevention of Cruelty to Animals Act 1986 and the Prevention of Cruelty to Animals Regulations 2008, and reviewed and permitted by the Melbourne University Animal Ethics Committee (AEEC ethics ID: 1413078).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
Comparative proteomic analysis of kinesin-8B deficient Plasmodium berghei during gametogenesis.J Proteomics. 2021 Mar 30;236:104118. doi: 10.1016/j.jprot.2021.104118. Epub 2021 Jan 21. J Proteomics. 2021. PMID: 33486016
-
Set regulation in asexual and sexual Plasmodium parasites reveals a novel mechanism of stage-specific expression.Mol Microbiol. 2006 May;60(4):870-82. doi: 10.1111/j.1365-2958.2006.05141.x. Mol Microbiol. 2006. PMID: 16677299
-
Functional Conservation of P48/45 Proteins in the Transmission Stages of Plasmodium vivax (Human Malaria Parasite) and P. berghei (Murine Malaria Parasite).mBio. 2018 Sep 4;9(5):e01627-18. doi: 10.1128/mBio.01627-18. mBio. 2018. PMID: 30181253 Free PMC article.
-
Asexual blood stages of malaria modulate gametocyte infectivity to the mosquito vector--possible implications for control strategies.Parasitology. 1991 Oct;103 Pt 2:191-6. doi: 10.1017/s0031182000059473. Parasitology. 1991. PMID: 1684037 Review.
-
Regulation of sexual commitment in malaria parasites - a complex affair.Curr Opin Microbiol. 2024 Jun;79:102469. doi: 10.1016/j.mib.2024.102469. Epub 2024 Apr 4. Curr Opin Microbiol. 2024. PMID: 38574448 Review.
Cited by
-
Coordinated regulation of gene expression in Plasmodium female gametocytes by two transcription factors.Elife. 2024 Jan 22;12:RP88317. doi: 10.7554/eLife.88317. Elife. 2024. PMID: 38252559 Free PMC article.
-
Zygote morphogenesis but not the establishment of cell polarity in Plasmodium berghei is controlled by the small GTPase, RAB11A.PLoS Pathog. 2020 May 28;16(5):e1008091. doi: 10.1371/journal.ppat.1008091. eCollection 2020 May. PLoS Pathog. 2020. PMID: 32463831 Free PMC article.
-
PfMFR3: A Multidrug-Resistant Modulator in Plasmodium falciparum.ACS Infect Dis. 2021 Apr 9;7(4):811-825. doi: 10.1021/acsinfecdis.0c00676. Epub 2021 Mar 14. ACS Infect Dis. 2021. PMID: 33715347 Free PMC article.
-
Alternative splicing is required for stage differentiation in malaria parasites.Genome Biol. 2019 Aug 1;20(1):151. doi: 10.1186/s13059-019-1756-6. Genome Biol. 2019. PMID: 31370870 Free PMC article.
-
Plasmodium berghei leucine-rich repeat protein 1 downregulates protein phosphatase 1 activity and is required for efficient oocyst development.Open Biol. 2022 Aug;12(8):220015. doi: 10.1098/rsob.220015. Epub 2022 Aug 3. Open Biol. 2022. PMID: 35920043 Free PMC article.
References
-
- Sinha A, Hughes KR, Modrzynska KK, Otto TD, Pfander C, Dickens NJ, Religa AA, Bushell E, Graham AL, Cameron R, Kafsack BFC, Williams AE, Llinás M, Berriman M, Billker O, Waters AP. A cascade of dna-binding proteins for sexual commitment and development in Plasmodium. Nature. 2014;507:253–7. doi: 10.1038/nature12970. - DOI - PMC - PubMed
-
- Janse CJ, Waters AP. Sexual development of parasites. In: Waters AP, Janse CJ, editors. Malaria Parasites: Genomes and Molecular Biology. Norfolk, England: Caister Academic Press; 2004.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

