Mitochondrial genomes reveal recombination in the presumed asexual Fusarium oxysporum species complex
- PMID: 28923029
- PMCID: PMC5604515
- DOI: 10.1186/s12864-017-4116-5
Mitochondrial genomes reveal recombination in the presumed asexual Fusarium oxysporum species complex
Abstract
Background: The Fusarium oxysporum species complex (FOSC) contains several phylogenetic lineages. Phylogenetic studies identified two to three major clades within the FOSC. The mitochondrial sequences are highly informative phylogenetic markers, but have been mostly neglected due to technical difficulties.
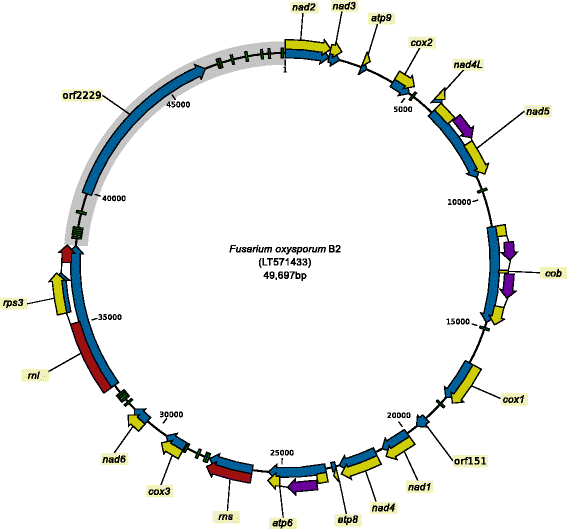
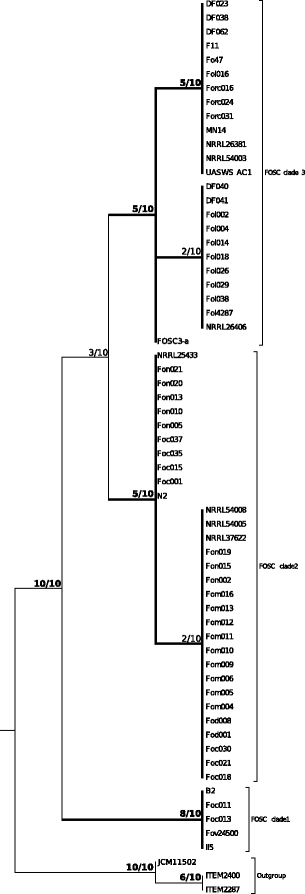
Results: A total of 61 complete mitogenomes of FOSC strains were de novo assembled and annotated. Length variations and intron patterns support the separation of three phylogenetic species. The variable region of the mitogenome that is typical for the genus Fusarium shows two new variants in the FOSC. The variant typical for Fusarium is found in members of all three clades, while variant 2 is found in clades 2 and 3 and variant 3 only in clade 2. The extended set of loci analyzed using a new implementation of the genealogical concordance species recognition method support the identification of three phylogenetic species within the FOSC. Comparative analysis of the mitogenomes in the FOSC revealed ongoing mitochondrial recombination within, but not between phylogenetic species.
Conclusions: The recombination indicates the presence of a parasexual cycle in F. oxysporum. The obstacles hindering the usage of the mitogenomes are resolved by using next generation sequencing and selective genome assemblers, such as GRAbB. Complete mitogenome sequences offer a stable basis and reference point for phylogenetic and population genetic studies.
Keywords: Comparative genomics; Mitochondrial genome; Mitochondrial recombination; Phylogenomics.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
-
- Baayen RP, O’Donnell K, Bonants PJM, Cigelnik E, Kroon LPNM, Roebroeck EJA, Waalwijk C. Gene genealogies and AFLP analyses in the Fusarium oxysporum complex identify monophyletic and nonmonophyletic formae speciales causing wilt and rot disease. Phytopathology. 2000;90(8):891–900. doi: 10.1094/PHYTO.2000.90.8.891. - DOI - PubMed
-
- O’Donnell K, Sutton DA, Wiederhold N, Robert VARG, Crous PW, Geiser DM. Veterinary Fusarioses within the United States. J Clin Microbiol (Epub ahead of print). 2016. doi:10.1128/JCM.01607-16. - DOI - PMC - PubMed
-
- van Diepeningen AD, Al-Hatmi AMS, Brankovics B, de Hoog GS. Taxonomy and Clinical Spectra of Fusarium Species: Where Do We Stand in 2014? Curr Clin Microbiol Rep. 2014;1(1-2):10–18. doi: 10.1007/s40588-014-0003-x. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

