DNA epigenome editing using CRISPR-Cas SunTag-directed DNMT3A
- PMID: 28923089
- PMCID: PMC5604343
- DOI: 10.1186/s13059-017-1306-z
DNA epigenome editing using CRISPR-Cas SunTag-directed DNMT3A
Abstract
Background: DNA methylation has widespread effects on gene expression during development. However, our ability to assign specific function to regions of DNA methylation is limited by the poor correlation between global patterns of DNA methylation and gene expression.
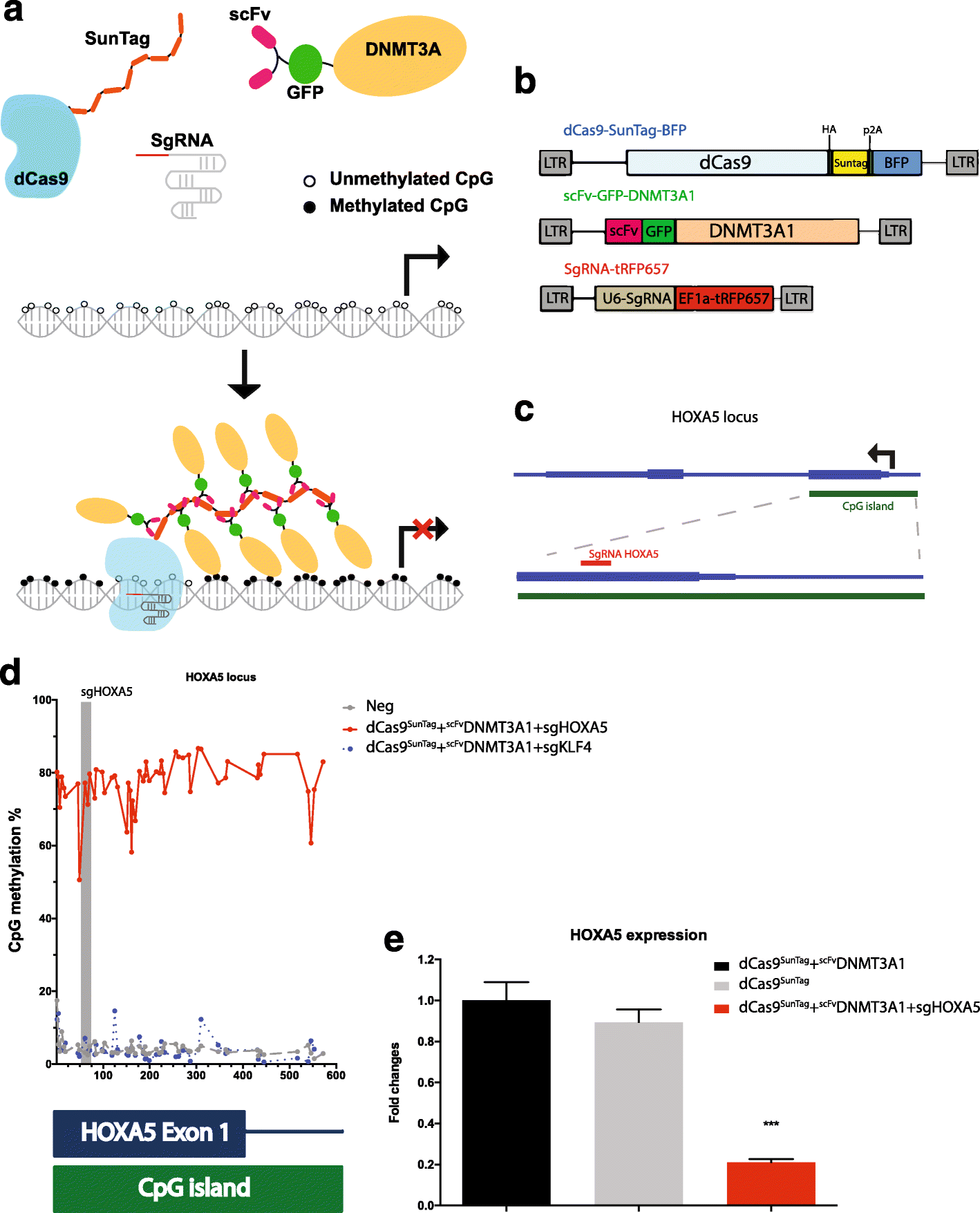
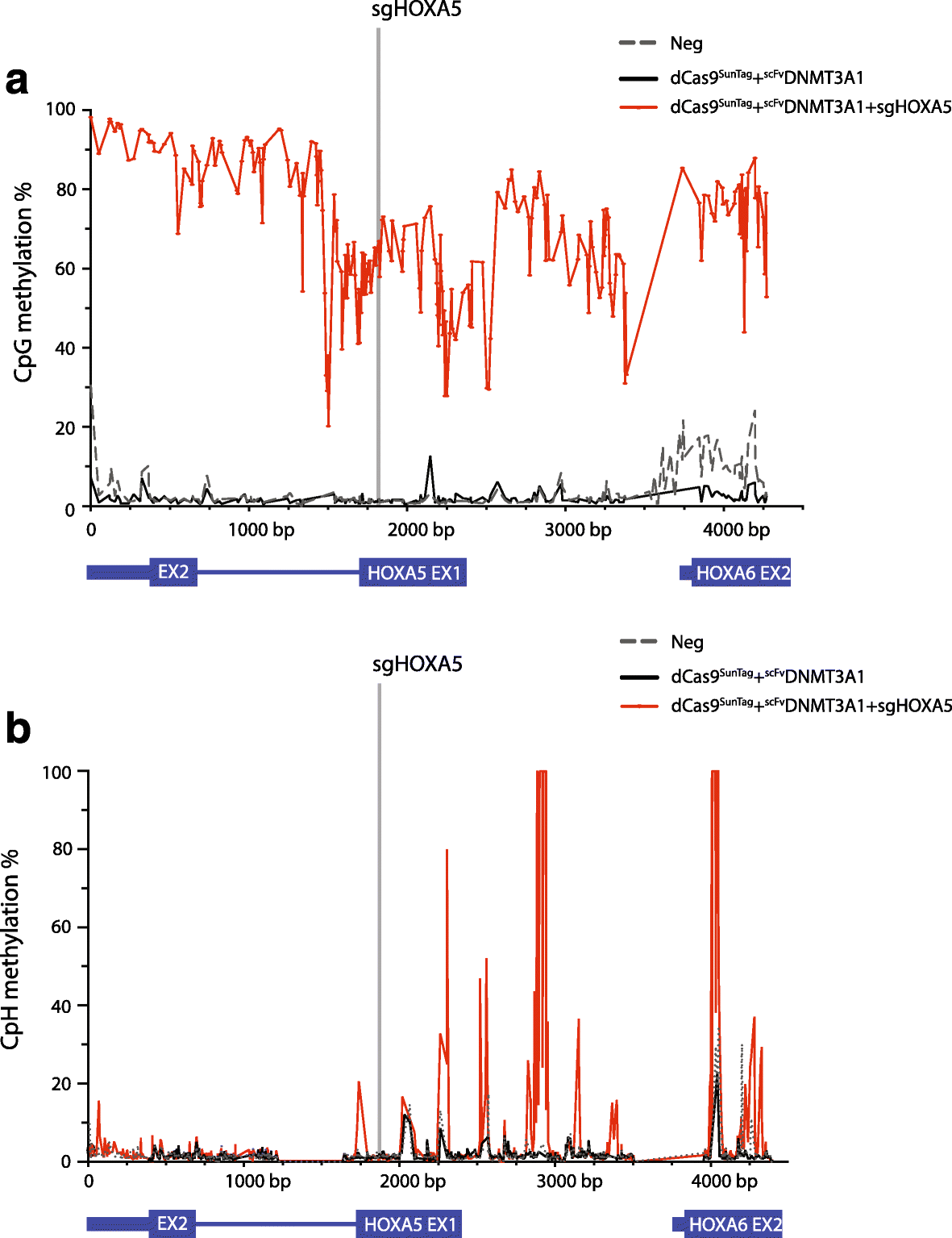
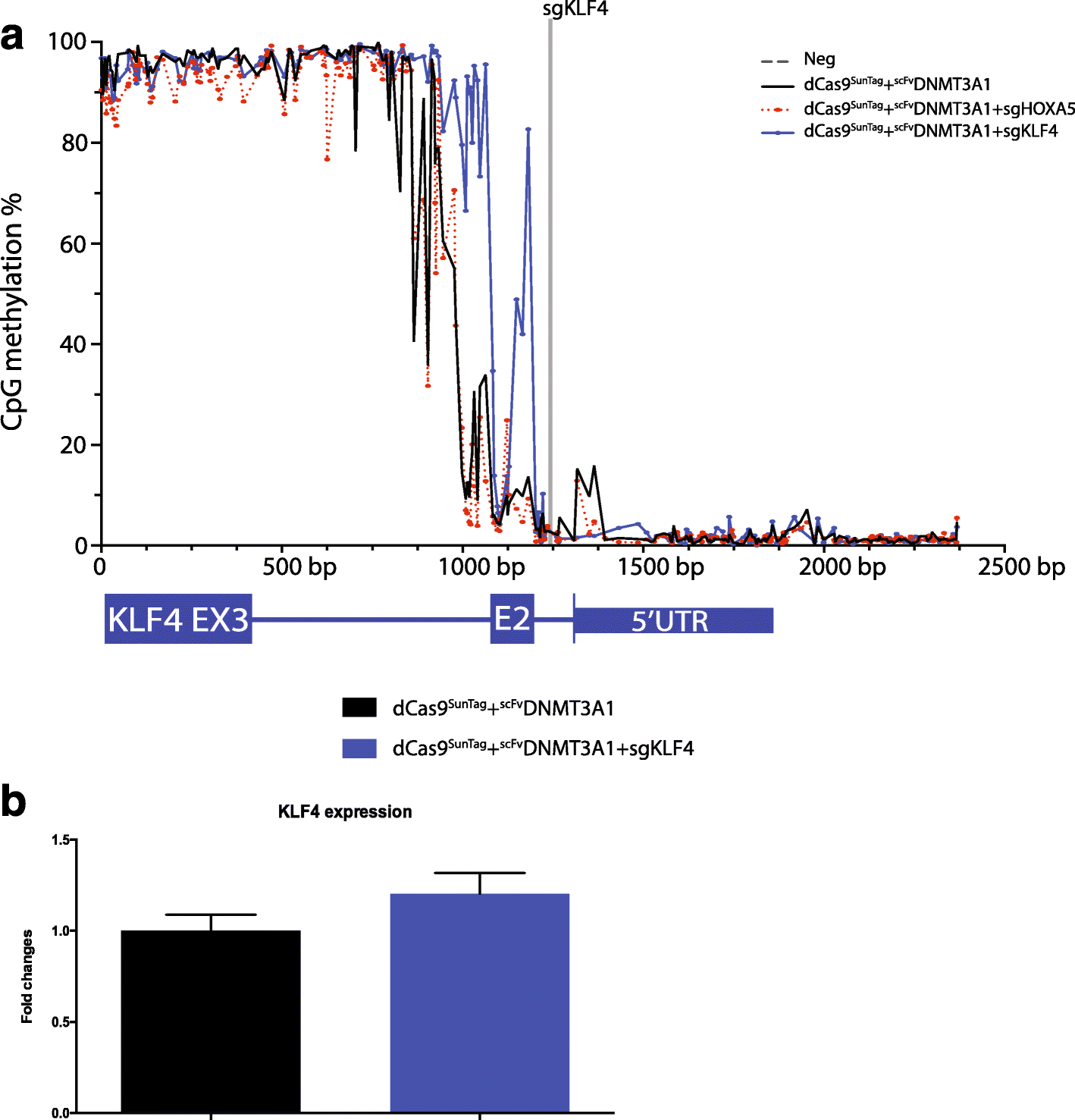
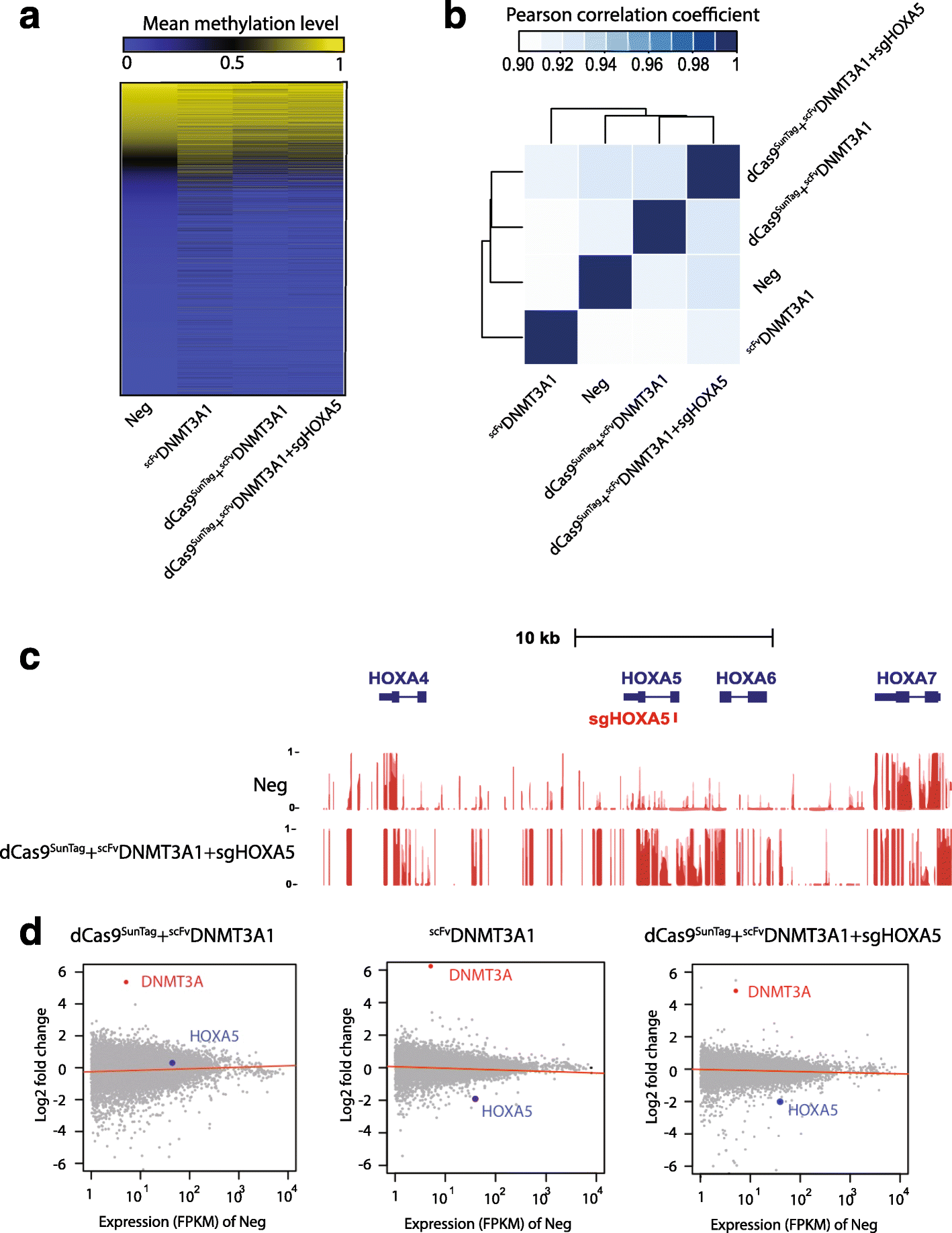
Results: Here, we utilize nuclease-deactivated Cas9 protein fused to repetitive peptide epitopes (SunTag) recruiting multiple copies of antibody-fused de novo DNA methyltransferase 3A (DNMT3A) (dCas9-SunTag-DNMT3A) to amplify the local DNMT3A concentration to methylate genomic sites of interest. We demonstrate that dCas9-SunTag-DNMT3A dramatically increases CpG methylation at the HOXA5 locus in human embryonic kidney (HEK293T) cells. Furthermore, using a single guide RNA, dCas9-SunTag-DNMT3A is able to methylate a 4.5-kb genomic region and repress HOXA5 gene expression. Reduced representation bisulfite sequencing and RNA-seq show that dCas9-SunTag-DNMT3A methylates regions of interest with minimal impact on the global DNA methylome and transcriptome.
Conclusions: This effective and precise tool enables site-specific manipulation of DNA methylation and may be used to address the relationship between DNA methylation and gene expression.
Conflict of interest statement
Ethics approval
Not applicable in this study.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
A modular dCas9-SunTag DNMT3A epigenome editing system overcomes pervasive off-target activity of direct fusion dCas9-DNMT3A constructs.Genome Res. 2018 Aug;28(8):1193-1206. doi: 10.1101/gr.233049.117. Epub 2018 Jun 15. Genome Res. 2018. PMID: 29907613 Free PMC article.
-
Characterization of Rationally Designed CRISPR/Cas9-Based DNA Methyltransferases with Distinct Methyltransferase and Gene Silencing Activities in Human Cell Lines and Primary Human T Cells.ACS Synth Biol. 2025 Feb 21;14(2):384-397. doi: 10.1021/acssynbio.4c00569. Epub 2025 Feb 3. ACS Synth Biol. 2025. PMID: 39898483
-
Efficient Targeted DNA Methylation with dCas9-Coupled DNMT3A-DNMT3L Methyltransferase.Methods Mol Biol. 2023;2577:177-188. doi: 10.1007/978-1-0716-2724-2_12. Methods Mol Biol. 2023. PMID: 36173573
-
A review on CRISPR/Cas-based epigenetic regulation in plants.Int J Biol Macromol. 2022 Oct 31;219:1261-1271. doi: 10.1016/j.ijbiomac.2022.08.182. Epub 2022 Aug 31. Int J Biol Macromol. 2022. PMID: 36057300 Review.
-
CRISPR/Cas-mediated macromolecular DNA methylation editing: Precision targeting of DNA methyltransferases in cancer therapy.Int J Biol Macromol. 2025 May;308(Pt 2):142401. doi: 10.1016/j.ijbiomac.2025.142401. Epub 2025 Mar 23. Int J Biol Macromol. 2025. PMID: 40132699 Review.
Cited by
-
In Vivo Hematopoietic Stem Cell Genome Editing: Perspectives and Limitations.Genes (Basel). 2022 Nov 27;13(12):2222. doi: 10.3390/genes13122222. Genes (Basel). 2022. PMID: 36553489 Free PMC article. Review.
-
Locus-Specific DNA Methylation Editing in Melanoma Cell Lines Using a CRISPR-Based System.Cancers (Basel). 2021 Oct 29;13(21):5433. doi: 10.3390/cancers13215433. Cancers (Basel). 2021. PMID: 34771597 Free PMC article.
-
Untapped Reserves: Controlling Primordial Follicle Growth Activation.Trends Mol Med. 2018 Mar;24(3):319-331. doi: 10.1016/j.molmed.2018.01.008. Epub 2018 Feb 13. Trends Mol Med. 2018. PMID: 29452791 Free PMC article. Review.
-
CRISPR technologies for precise epigenome editing.Nat Cell Biol. 2021 Jan;23(1):11-22. doi: 10.1038/s41556-020-00620-7. Epub 2021 Jan 8. Nat Cell Biol. 2021. PMID: 33420494 Review.
-
Stability and DNA Methyltransferase Activity of DNMT3A are Maintained by Ubiquitin-Specific Peptidase 11 (USP11) and Sumoylation Countering Degradation.bioRxiv [Preprint]. 2025 Jun 27:2025.03.05.641683. doi: 10.1101/2025.03.05.641683. bioRxiv. 2025. PMID: 40161590 Free PMC article. Preprint.
References
MeSH terms
Substances
Grants and funding
- R01 CA193466/CA/NCI NIH HHS/United States
- F99 CA222736/CA/NCI NIH HHS/United States
- R56 DK092883/DK/NIDDK NIH HHS/United States
- R01 CA193235/CA/NCI NIH HHS/United States
- P50 CA126752/CA/NCI NIH HHS/United States
- P30 CA125123/CA/NCI NIH HHS/United States
- K00 CA222736/CA/NCI NIH HHS/United States
- P30 AI036211/AI/NIAID NIH HHS/United States
- S10 RR024574/RR/NCRR NIH HHS/United States
- T32 DK060445/DK/NIDDK NIH HHS/United States
- R01 DK092883/DK/NIDDK NIH HHS/United States
- R01 CA183252/CA/NCI NIH HHS/United States
- R01 HG007538/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

