Interaction networks, ecological stability, and collective antibiotic tolerance in polymicrobial infections
- PMID: 28923953
- PMCID: PMC5635929
- DOI: 10.1073/pnas.1713372114
Interaction networks, ecological stability, and collective antibiotic tolerance in polymicrobial infections
Abstract
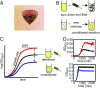
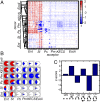
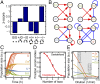
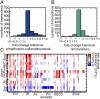
Polymicrobial infections constitute small ecosystems that accommodate several bacterial species. Commonly, these bacteria are investigated in isolation. However, it is unknown to what extent the isolates interact and whether their interactions alter bacterial growth and ecosystem resilience in the presence and absence of antibiotics. We quantified the complete ecological interaction network for 72 bacterial isolates collected from 23 individuals diagnosed with polymicrobial urinary tract infections and found that most interactions cluster based on evolutionary relatedness. Statistical network analysis revealed that competitive and cooperative reciprocal interactions are enriched in the global network, while cooperative interactions are depleted in the individual host community networks. A population dynamics model parameterized by our measurements suggests that interactions restrict community stability, explaining the observed species diversity of these communities. We further show that the clinical isolates frequently protect each other from clinically relevant antibiotics. Together, these results highlight that ecological interactions are crucial for the growth and survival of bacteria in polymicrobial infection communities and affect their assembly and resilience.
Keywords: antibiotics; infection; microbiology; systems biology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

