Prepatterning by RhoGEFs governs Rho GTPase spatiotemporal dynamics during wound repair
- PMID: 28923977
- PMCID: PMC5716286
- DOI: 10.1083/jcb.201704145
Prepatterning by RhoGEFs governs Rho GTPase spatiotemporal dynamics during wound repair
Abstract
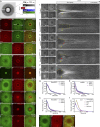
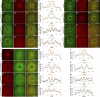
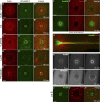
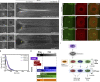
Like tissues, single cells are subjected to continual stresses and damage. As such, cells have a robust wound repair mechanism comprised of dynamic membrane resealing and cortical cytoskeletal remodeling. One group of proteins, the Rho family of small guanosine triphosphatases (GTPases), is critical for this actin and myosin cytoskeletal response in which they form distinct dynamic spatial and temporal patterns/arrays surrounding the wound. A key mechanistic question, then, is how these GTPase arrays are formed. Here, we show that in the Drosophila melanogaster cell wound repair model Rho GTPase arrays form in response to prepatterning by Rho guanine nucleotide exchange factors (RhoGEFs), a family of proteins involved in the activation of small GTPases. Furthermore, we show that Annexin B9, a member of a class of proteins associated with the membrane resealing, is involved in an early, Rho family-independent, actin stabilization that is integral to the formation of one RhoGEF array. Thus, Annexin proteins may link membrane resealing to cytoskeletal remodeling processes in single cell wound repair.
© 2017 Nakamura et al.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

