Pan-genomic analyses identify key Helicobacter pylori pathogenic loci modified by carcinogenic host microenvironments
- PMID: 28924022
- PMCID: PMC5857411
- DOI: 10.1136/gutjnl-2017-313863
Pan-genomic analyses identify key Helicobacter pylori pathogenic loci modified by carcinogenic host microenvironments
Abstract
Objective: Helicobacter pylori is the strongest risk factor for gastric cancer; however, the majority of infected individuals do not develop disease. Pathological outcomes are mediated by complex interactions among bacterial, host and environmental constituents, and two dietary factors linked with gastric cancer risk are iron deficiency and high salt. We hypothesised that prolonged adaptation of H. pylori to in vivo carcinogenic microenvironments results in genetic modification important for disease.
Design: Whole genome sequencing of genetically related H. pylori strains that differ in virulence and targeted H. pylori sequencing following prolonged exposure of bacteria to in vitro carcinogenic conditions were performed.
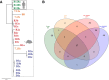
Results: A total of 180 unique single nucleotide polymorphisms (SNPs) were identified among the collective genomes when compared with a reference H. pylori genome. Importantly, common SNPs were identified in isolates harvested from iron-depleted and high salt carcinogenic microenvironments, including an SNP within fur (FurR88H). To investigate the direct role of low iron and/or high salt, H. pylori was continuously cultured in vitro under low iron or high salt conditions to assess fur genetic variation. Exposure to low iron or high salt selected for the FurR88H variant after only 5 days. To extend these results, fur was sequenced in 339 clinical H. pylori strains. Among the isolates examined, 17% (40/232) of strains isolated from patients with premalignant lesions harboured the FurR88H variant, compared with only 6% (6/107) of strains from patients with non-atrophic gastritis alone (p=0.0034).
Conclusion: These results indicate that specific genetic variation arises within H. pylori strains during in vivo adaptation to conditions conducive for gastric carcinogenesis.
Keywords: Helicobacter pylori; gastric cancer; high salt; iron deficiency.
© Article author(s) (or their employer(s) unless otherwise stated in the text of the article) 2018. All rights reserved. No commercial use is permitted unless otherwise expressly granted.
Conflict of interest statement
Competing interests: None declared.
Figures



Similar articles
-
Genetic Manipulation of Helicobacter pylori Virulence Function by Host Carcinogenic Phenotypes.Cancer Res. 2017 May 1;77(9):2401-2412. doi: 10.1158/0008-5472.CAN-16-2922. Epub 2017 Feb 16. Cancer Res. 2017. PMID: 28209611 Free PMC article.
-
Iron deficiency accelerates Helicobacter pylori-induced carcinogenesis in rodents and humans.J Clin Invest. 2013 Jan;123(1):479-92. doi: 10.1172/JCI64373. Epub 2012 Dec 21. J Clin Invest. 2013. PMID: 23257361 Free PMC article. Clinical Trial.
-
Host genetic factors respond to pathogenic step-specific virulence factors of Helicobacter pylori in gastric carcinogenesis.Mutat Res Rev Mutat Res. 2014 Jan-Mar;759:14-26. doi: 10.1016/j.mrrev.2013.09.002. Epub 2013 Sep 25. Mutat Res Rev Mutat Res. 2014. PMID: 24076409 Review.
-
SNP interactions of Helicobacter pylori-related host genes PGC, PTPN11, IL1B, and TLR4 in susceptibility to gastric carcinogenesis.Oncotarget. 2015 Aug 7;6(22):19017-26. doi: 10.18632/oncotarget.4231. Oncotarget. 2015. PMID: 26158864 Free PMC article.
-
Genetic host factors in Helicobacter pylori-induced carcinogenesis: Emerging new paradigms.Biochim Biophys Acta Rev Cancer. 2018 Jan;1869(1):42-52. doi: 10.1016/j.bbcan.2017.11.003. Epub 2017 Nov 24. Biochim Biophys Acta Rev Cancer. 2018. PMID: 29154808 Review.
Cited by
-
Positive Selection of Mutations in the Helicobacter pylori katA 5' Untranslated Region in a Mongolian Gerbil Model of Gastric Disease.Infect Immun. 2022 Jul 21;90(7):e0000422. doi: 10.1128/iai.00004-22. Epub 2022 Jun 2. Infect Immun. 2022. PMID: 35652648 Free PMC article.
-
Review of current diagnostic methods and advances in Helicobacter pylori diagnostics in the era of next generation sequencing.World J Gastroenterol. 2019 Aug 28;25(32):4629-4660. doi: 10.3748/wjg.v25.i32.4629. World J Gastroenterol. 2019. PMID: 31528091 Free PMC article. Review.
-
Genomic diversity of Helicobacter pylori populations from different regions of the human stomach.Gut Microbes. 2022 Jan-Dec;14(1):2152306. doi: 10.1080/19490976.2022.2152306. Gut Microbes. 2022. PMID: 36469575 Free PMC article.
-
Maturation of atypical ribosomal RNA precursors in Helicobacter pylori.Nucleic Acids Res. 2019 Jun 20;47(11):5906-5921. doi: 10.1093/nar/gkz258. Nucleic Acids Res. 2019. PMID: 31006803 Free PMC article.
-
Genetics of Host Protection against Helicobacter pylori Infections.Int J Mol Sci. 2021 Mar 21;22(6):3192. doi: 10.3390/ijms22063192. Int J Mol Sci. 2021. PMID: 33801073 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
