DNA Methylomes Reveal Biological Networks Involved in Human Eye Development, Functions and Associated Disorders
- PMID: 28924151
- PMCID: PMC5603607
- DOI: 10.1038/s41598-017-12084-1
DNA Methylomes Reveal Biological Networks Involved in Human Eye Development, Functions and Associated Disorders
Abstract
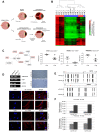
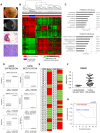
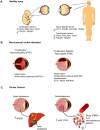
This work provides a comprehensive CpG methylation landscape of the different layers of the human eye that unveils the gene networks associated with their biological functions and how these are disrupted in common visual disorders. Herein, we firstly determined the role of CpG methylation in the regulation of ocular tissue-specification and described hypermethylation of retinal transcription factors (i.e., PAX6, RAX, SIX6) in a tissue-dependent manner. Second, we have characterized the DNA methylome of visual disorders linked to internal and external environmental factors. Main conclusions allow certifying that crucial pathways related to Wnt-MAPK signaling pathways or neuroinflammation are epigenetically controlled in the fibrotic disorders involved in retinal detachment, but results also reinforced the contribution of neurovascularization (ETS1, HES5, PRDM16) in diabetic retinopathy. Finally, we had studied the methylome in the most frequent intraocular tumors in adults and children (uveal melanoma and retinoblastoma, respectively). We observed that hypermethylation of tumor suppressor genes is a frequent event in ocular tumors, but also unmethylation is associated with tumorogenesis. Interestingly, unmethylation of the proto-oncogen RAB31 was a predictor of metastasis risk in uveal melanoma. Loss of methylation of the oncogenic mir-17-92 cluster was detected in primary tissues but also in blood from patients.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

