Genetic Code Optimization for Cotranslational Protein Folding: Codon Directional Asymmetry Correlates with Antiparallel Betasheets, tRNA Synthetase Classes
- PMID: 28924459
- PMCID: PMC5591391
- DOI: 10.1016/j.csbj.2017.08.001
Genetic Code Optimization for Cotranslational Protein Folding: Codon Directional Asymmetry Correlates with Antiparallel Betasheets, tRNA Synthetase Classes
Abstract
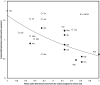
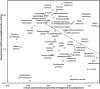
A new codon property, codon directional asymmetry in nucleotide content (CDA), reveals a biologically meaningful genetic code dimension: palindromic codons (first and last nucleotides identical, codon structure XZX) are symmetric (CDA = 0), codons with structures ZXX/XXZ are 5'/3' asymmetric (CDA = - 1/1; CDA = - 0.5/0.5 if Z and X are both purines or both pyrimidines, assigning negative/positive (-/+) signs is an arbitrary convention). Negative/positive CDAs associate with (a) Fujimoto's tetrahedral codon stereo-table; (b) tRNA synthetase class I/II (aminoacylate the 2'/3' hydroxyl group of the tRNA's last ribose, respectively); and (c) high/low antiparallel (not parallel) betasheet conformation parameters. Preliminary results suggest CDA-whole organism associations (body temperature, developmental stability, lifespan). Presumably, CDA impacts spatial kinetics of codon-anticodon interactions, affecting cotranslational protein folding. Some synonymous codons have opposite CDA sign (alanine, leucine, serine, and valine), putatively explaining how synonymous mutations sometimes affect protein function. Correlations between CDA and tRNA synthetase classes are weaker than between CDA and antiparallel betasheet conformation parameters. This effect is stronger for mitochondrial genetic codes, and potentially drives mitochondrial codon-amino acid reassignments. CDA reveals information ruling nucleotide-protein relations embedded in reversed (not reverse-complement) sequences (5'-ZXX-3'/5'-XXZ-3').
Keywords: Alpha helix; Beta turn; Codon-amino acid assignment; Mitochondrial genetic code; Secondary structure; Synonymous codon.
Figures




Similar articles
-
Codon Directional Asymmetry Suggests Swapped Prebiotic 1st and 2nd Codon Positions.Int J Mol Sci. 2020 Jan 5;21(1):347. doi: 10.3390/ijms21010347. Int J Mol Sci. 2020. PMID: 31948054 Free PMC article.
-
Bias for 3'-Dominant Codon Directional Asymmetry in Theoretical Minimal RNA Rings.J Comput Biol. 2019 Sep;26(9):1003-1012. doi: 10.1089/cmb.2018.0256. Epub 2019 May 22. J Comput Biol. 2019. PMID: 31120344
-
Evolution of the genetic code through progressive symmetry breaking.J Theor Biol. 2014 Apr 21;347:95-108. doi: 10.1016/j.jtbi.2014.01.002. Epub 2014 Jan 14. J Theor Biol. 2014. PMID: 24434741
-
The Importance of Being Modified: The Role of RNA Modifications in Translational Fidelity.Enzymes. 2017;41:1-50. doi: 10.1016/bs.enz.2017.03.005. Epub 2017 Apr 22. Enzymes. 2017. PMID: 28601219 Free PMC article. Review.
-
Genetic code 1990. Outlook.Experientia. 1990 Dec 1;46(11-12):1149-57. doi: 10.1007/BF01936925. Experientia. 1990. PMID: 2147658 Review.
Cited by
-
Network analysis of synonymous codon usage.Bioinformatics. 2020 Dec 8;36(19):4876-4884. doi: 10.1093/bioinformatics/btaa603. Bioinformatics. 2020. PMID: 32609328 Free PMC article.
-
Genome Evolution from Random Ligation of RNAs of Autocatalytic Sets.Int J Mol Sci. 2021 Dec 16;22(24):13526. doi: 10.3390/ijms222413526. Int J Mol Sci. 2021. PMID: 34948321 Free PMC article.
-
RNA Rings Strengthen Hairpin Accretion Hypotheses for tRNA Evolution: A Reply to Commentaries by Z.F. Burton and M. Di Giulio.J Mol Evol. 2020 Apr;88(3):243-252. doi: 10.1007/s00239-020-09929-1. Epub 2020 Feb 5. J Mol Evol. 2020. PMID: 32025759
-
Bijective codon transformations show genetic code symmetries centered on cytosine's coding properties.Theory Biosci. 2018 Apr;137(1):17-31. doi: 10.1007/s12064-017-0258-x. Epub 2017 Nov 16. Theory Biosci. 2018. PMID: 29147851
-
Pentamers with Non-redundant Frames: Bias for Natural Circular Code Codons.J Mol Evol. 2020 Mar;88(2):194-201. doi: 10.1007/s00239-019-09925-0. Epub 2020 Jan 7. J Mol Evol. 2020. PMID: 31907555
References
-
- Di Giulio M. The extension reached by the minimization of the polarity distances during the evolution of the genetic code. J Mol Evol. 1989;29:288–293. - PubMed
-
- Haig D., Hurst L.D. A quantitative measure of error minimization in the genetic-code. J Mol Evol. 1991;33:412–417. - PubMed
-
- Ardell D.H. On error minimization in a sequential origin of the standard genetic code. J Mol Evol. 1998;47:1–13. - PubMed
-
- Freeland S.J., Hurst L.D. Load minimization of the genetic code: history does not explain the pattern. Proc R Soc B Biol Sci. 1998;265:2111–2119.
LinkOut - more resources
Full Text Sources
Other Literature Sources
