Biochemical and structural characterization of a novel arginine kinase from the spider Polybetes pythagoricus
- PMID: 28924503
- PMCID: PMC5598448
- DOI: 10.7717/peerj.3787
Biochemical and structural characterization of a novel arginine kinase from the spider Polybetes pythagoricus
Abstract
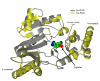
Energy buffering systems are key for homeostasis during variations in energy supply. Spiders are the most important predators for insects and therefore key in terrestrial ecosystems. From biomedical interest, spiders are important for their venoms and as a source of potent allergens, such as arginine kinase (AK, EC 2.7.3.3). AK is an enzyme crucial for energy metabolism, keeping the pool of phosphagens in invertebrates, and also an allergen for humans. In this work, we studied AK from the Argentininan spider Polybetes pythagoricus (PpAK), from its complementary DNA to the crystal structure. The PpAK cDNA from muscle was cloned, and it is comprised of 1068 nucleotides that encode a 384-amino acids protein, similar to other invertebrate AKs. The apparent Michaelis-Menten kinetic constant (Km ) was 1.7 mM with a kcat of 75 s-1. Two crystal structures are presented, the apoPvAK and PpAK bound to arginine, both in the open conformation with the active site lid (residues 310-320) completely disordered. The guanidino group binding site in the apo structure appears to be organized to accept the arginine substrate. Finally, these results contribute to knowledge of mechanistic details of the function of arginine kinase.
Keywords: Allergen; Arachnida; Arginine kinase; Arthropoda; Crystal structure; Open conformation; Phosphagen; Polybetes pytagoricus; Spider; cDNA cloning.
Conflict of interest statement
The author declares there are no competing interests.
Figures

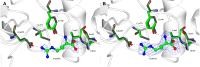
Similar articles
-
Arginine kinase evolved twice: evidence that echinoderm arginine kinase originated from creatine kinase.Biochem J. 1999 Jun 15;340 ( Pt 3)(Pt 3):671-5. Biochem J. 1999. PMID: 10359650 Free PMC article.
-
Characterization of phenoloxidase activity from spider Polybetes pythagoricus hemocyanin.J Exp Zool A Ecol Genet Physiol. 2015 Oct;323(8):547-55. doi: 10.1002/jez.1947. Epub 2015 Jul 14. J Exp Zool A Ecol Genet Physiol. 2015. PMID: 26173645
-
A novel arginine kinase from the shrimp Neocaridina denticulata: the fourth arginine kinase gene lineage.Gene. 2009 May 15;437(1-2):80-7. doi: 10.1016/j.gene.2009.02.018. Epub 2009 Mar 5. Gene. 2009. PMID: 19268694
-
Toxocara canis: molecular cloning, characterization, expression and comparison of the kinetics of cDNA-derived arginine kinase.Exp Parasitol. 2007 Oct;117(2):124-32. doi: 10.1016/j.exppara.2007.03.015. Epub 2007 Apr 4. Exp Parasitol. 2007. PMID: 17574244
-
Evolution and physiological roles of phosphagen systems.Annu Rev Physiol. 2001;63:289-325. doi: 10.1146/annurev.physiol.63.1.289. Annu Rev Physiol. 2001. PMID: 11181958 Review.
Cited by
-
A comprehensive review of arginine kinase proteins: What we need to know?Biochem Biophys Rep. 2024 Oct 8;40:101837. doi: 10.1016/j.bbrep.2024.101837. eCollection 2024 Dec. Biochem Biophys Rep. 2024. PMID: 39435382 Free PMC article. Review.
-
Crystal Structure of H227A Mutant of Arginine Kinase in Daphnia magna Suggests the Importance of Its Stability.Molecules. 2022 Jan 28;27(3):884. doi: 10.3390/molecules27030884. Molecules. 2022. PMID: 35164149 Free PMC article.
-
Natural Products Containing 'Rare' Organophosphorus Functional Groups.Molecules. 2019 Feb 28;24(5):866. doi: 10.3390/molecules24050866. Molecules. 2019. PMID: 30823503 Free PMC article. Review.
-
Chemical profiling and arginine kinase inhibitory activity of Angelica dahurica leaves.Heliyon. 2024 Mar 10;10(6):e27589. doi: 10.1016/j.heliyon.2024.e27589. eCollection 2024 Mar 30. Heliyon. 2024. PMID: 38509962 Free PMC article.
-
Binding of green tea epigallocatechin gallate to the arginine kinase active site from the brown recluse spider (Loxosceles laeta): A potential synergist to chemical pesticides.Heliyon. 2024 Jul 3;10(13):e34036. doi: 10.1016/j.heliyon.2024.e34036. eCollection 2024 Jul 15. Heliyon. 2024. PMID: 39071691 Free PMC article.
References
-
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallographica Section D. 2010;66:213–221. doi: 10.1107/S0907444909052925. - DOI - PMC - PubMed
-
- Arjunwadkar AV, Raghupathi Rami Reddy S. Characterization and distribution of arginine kinase in the tissues of the scorpion, Palamneus phipsoni. Canadian Journal of Zoology. 1985;63:2262–2266. doi: 10.1139/z85-335. - DOI
-
- Ausubel FM, Brent R, Kingston R, Moore D, Seidman J, Smith J, Sthruhl K. Short protocols in molecular biology. John Wiley & Sons; New York: 1995.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

