It's fun to transcribe with Fun30: A model for nucleosome dynamics during RNA polymerase II-mediated elongation
- PMID: 28925811
- PMCID: PMC5834224
- DOI: 10.1080/21541264.2017.1356434
It's fun to transcribe with Fun30: A model for nucleosome dynamics during RNA polymerase II-mediated elongation
Abstract
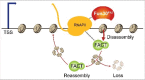
The ability of elongating RNA polymerase II (RNAPII) to regulate the nucleosome barrier is poorly understood because we do not know enough about the involved factors and we lack a conceptual framework to model this process. Our group recently identified the conserved Fun30/SMARCAD1 family chromatin-remodeling factor, Fun30Fft3, as being critical for relieving the nucleosome barrier during RNAPII-mediated elongation, and proposed a model illustrating how Fun30Fft3 may contribute to nucleosome disassembly during RNAPII-mediated elongation. Here, we present a model that describes nucleosome dynamics during RNAPII-mediated elongation in mathematical terms and addresses the involvement of Fun30Fft3 in this process.
Keywords: Fun30; RNAPII elongation; chromatin remodeler; histone chaperone FACT; nucleosome barrier; nucleosome disassembly.
Figures
Similar articles
-
Chromatin remodeller Fun30Fft3 induces nucleosome disassembly to facilitate RNA polymerase II elongation.Nat Commun. 2017 Feb 20;8:14527. doi: 10.1038/ncomms14527. Nat Commun. 2017. PMID: 28218250 Free PMC article.
-
Spt4 facilitates the movement of RNA polymerase II through the +2 nucleosomal barrier.Cell Rep. 2021 Sep 28;36(13):109755. doi: 10.1016/j.celrep.2021.109755. Cell Rep. 2021. PMID: 34592154 Free PMC article.
-
To cross or not to cross the nucleosome, that is the elongation question..RNA Biol. 2011 May-Jun;8(3):389-93. doi: 10.4161/rna.8.3.14334. Epub 2011 May 1. RNA Biol. 2011. PMID: 21445002
-
Negotiating the nucleosome: factors that allow RNA polymerase II to elongate through chromatin.Biochem Cell Biol. 2007 Aug;85(4):426-34. doi: 10.1139/O07-054. Biochem Cell Biol. 2007. PMID: 17713578 Review.
-
A conserved role of the RSC chromatin remodeler in the establishment of nucleosome-depleted regions.Curr Genet. 2017 May;63(2):187-193. doi: 10.1007/s00294-016-0642-y. Epub 2016 Aug 24. Curr Genet. 2017. PMID: 27558480 Free PMC article. Review.
Cited by
-
Fun30 and Rtt109 Mediate Epigenetic Regulation of the DNA Damage Response Pathway in C. albicans.J Fungi (Basel). 2022 May 25;8(6):559. doi: 10.3390/jof8060559. J Fungi (Basel). 2022. PMID: 35736042 Free PMC article.
-
Nucleosome Remodeling by Fun30SMARCAD1 in the DNA Damage Response.Front Mol Biosci. 2019 Sep 4;6:78. doi: 10.3389/fmolb.2019.00078. eCollection 2019. Front Mol Biosci. 2019. PMID: 31555662 Free PMC article. Review.
-
Fun30 nucleosome remodeller regulates white-to-opaque switching in Candida albicans.Acta Biochim Biophys Sin (Shanghai). 2023 Mar 25;55(3):508-517. doi: 10.3724/abbs.2023031. Acta Biochim Biophys Sin (Shanghai). 2023. PMID: 36896644 Free PMC article.
References
-
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251-60. http://doi.org/10.1038/38444. PMID:9305837 - DOI - PubMed
-
- Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423:145-50. http://doi.org/10.1038/nature01595. PMID:12736678 - DOI - PubMed
-
- Lorch Y, LaPointe JW, Kornberg RD. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell. 1987;49:203-10. http://doi.org/10.1016/0092-8674(87)90561-7. PMID:3568125 - DOI - PubMed
-
- Izban MG, Luse DS. Transcription on nucleosomal templates by RNA polymerase II in vitro: inhibition of elongation with enhancement of sequence-specific pausing. Genes Dev. 1991;5:683-96. http://doi.org/10.1101/gad.5.4.683 - DOI - PubMed
-
- Knezetic JA, Luse DS. The presence of nucleosomes on a DNA template prevents initiation by RNA polymerase II in vitro. Cell. 1986;45:95-104. http://doi.org/10.1016/0092-8674(86)90541-6. PMID:3955658 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
