Loss of RUNX1 is associated with aggressive lung adenocarcinomas
- PMID: 28926105
- PMCID: PMC5989135
- DOI: 10.1002/jcp.26201
Loss of RUNX1 is associated with aggressive lung adenocarcinomas
Abstract
The mammalian runt-related factor 1 (RUNX1) is a master transcription factor that regulates lineage specification of hematopoietic stem cells. RUNX1 translocations result in the development of myeloid leukemias. Recently, RUNX1 has been implicated as a tumor suppressor in other cancers. We postulated RUNX1 expression may be associated with lung adenocarcinoma etiology and/or progression. We evaluated the association of RUNX1 mRNA expression with overall survival data from The Cancer Genome Atlas (TCGA), a publically available database. Compared to high expression levels, Low RUNX1 levels from lung adenocarcinomas were associated with a worse overall survival (Hazard Ratio = 2.014 (1.042-3.730 95% confidence interval), log-rank p = 0.035) compared to those that expressed high RUNX1 levels. Further immunohistochemical examination of 85 surgical specimens resected at the University of Vermont Medical Center identified that low RUNX1 protein expression was associated with larger tumors (p = 0.038). Gene expression network analysis was performed on the same subset of TCGA cases that demonstrated differential survival by RUNX1 expression. This analysis, which reveals regulatory relationships, showed that reduced RUNX1 levels were closely linked to upregulation of the transcription factor E2F1. To interrogate this relationship, RUNX1 was depleted in a lung cancer cell line that expresses high levels of RUNX1. Loss of RUNX1 resulted in enhanced proliferation, migration, and invasion. RUNX1 depletion also resulted in increased mRNA expression of E2F1 and multiple E2F1 target genes. Our data implicate loss of RUNX1 as driver of lung adenocarcinoma aggression, potentially through deregulation of the E2F1 pathway.
Keywords: RUNX; adenocarcinoma; non-small cell lung cancer; runt related transcription factor.
© 2017 Wiley Periodicals, Inc.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures



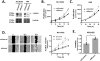
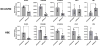

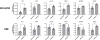
Similar articles
-
RUNX1-dependent mechanisms in biological control and dysregulation in cancer.J Cell Physiol. 2019 Jun;234(6):8597-8609. doi: 10.1002/jcp.27841. Epub 2018 Dec 4. J Cell Physiol. 2019. PMID: 30515788 Free PMC article. Review.
-
Transcriptional Auto-Regulation of RUNX1 P1 Promoter.PLoS One. 2016 Feb 22;11(2):e0149119. doi: 10.1371/journal.pone.0149119. eCollection 2016. PLoS One. 2016. PMID: 26901859 Free PMC article.
-
RUNX1 promotes proliferation and migration in non-small cell lung cancer cell lines via the mTOR pathway.FASEB J. 2023 Nov;37(11):e23195. doi: 10.1096/fj.202300687RR. FASEB J. 2023. PMID: 37801076
-
RUNX1 predicts poor prognosis and correlates with tumor progression in clear cell renal carcinoma.Pathol Res Pract. 2023 Nov;251:154886. doi: 10.1016/j.prp.2023.154886. Epub 2023 Oct 12. Pathol Res Pract. 2023. PMID: 37844486
-
A role for RUNX1 in hematopoiesis and myeloid leukemia.Int J Hematol. 2013 Jun;97(6):726-34. doi: 10.1007/s12185-013-1347-3. Epub 2013 Apr 24. Int J Hematol. 2013. PMID: 23613270 Review.
Cited by
-
RUNX transcription factors: biological functions and implications in cancer.Clin Exp Med. 2024 Mar 2;24(1):50. doi: 10.1007/s10238-023-01281-0. Clin Exp Med. 2024. PMID: 38430423 Free PMC article. Review.
-
Signatures of Co-Deregulated Genes and Their Transcriptional Regulators in Lung Cancer.Int J Mol Sci. 2022 Sep 18;23(18):10933. doi: 10.3390/ijms231810933. Int J Mol Sci. 2022. PMID: 36142846 Free PMC article.
-
Specific Deletions of Chromosomes 3p, 5q, 13q, and 21q among Patients with G2 Grade of Non-Small Cell Lung Cancer.Int J Mol Sci. 2024 Aug 8;25(16):8642. doi: 10.3390/ijms25168642. Int J Mol Sci. 2024. PMID: 39201328 Free PMC article.
-
LINC02126 is a potential diagnostic, prognostic and immunotherapeutic target for lung adenocarcinoma.BMC Pulm Med. 2022 Nov 10;22(1):412. doi: 10.1186/s12890-022-02215-4. BMC Pulm Med. 2022. PMID: 36357869 Free PMC article.
-
RUNX1-dependent mechanisms in biological control and dysregulation in cancer.J Cell Physiol. 2019 Jun;234(6):8597-8609. doi: 10.1002/jcp.27841. Epub 2018 Dec 4. J Cell Physiol. 2019. PMID: 30515788 Free PMC article. Review.
References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. - PubMed
-
- Barnes GL, et al. Osteoblast-related transcription factors Runx2 (Cbfa1/AML3) and MSX2 mediate the expression of bone sialoprotein in human metastatic breast cancer cells. Cancer Res. 2003;63(10):2631–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

