The microbiota of water buffalo milk during mastitis
- PMID: 28926595
- PMCID: PMC5604978
- DOI: 10.1371/journal.pone.0184710
The microbiota of water buffalo milk during mastitis
Abstract
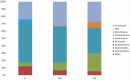
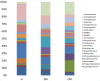
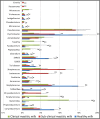
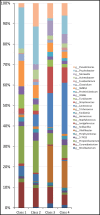
The aim of this study was to define the microbiota of water buffalo milk during sub-clinical and clinical mastitis, as compared to healthy status, by using high-throughput sequencing of the 16S rRNA gene. A total of 137 quarter samples were included in the experimental design: 27 samples derived from healthy, culture negative quarters, with a Somatic Cell Count (SCC) of less than 200,000 cells/ml; 27 samples from quarters with clinical mastitis; 83 samples were collected from quarters with subclinical mastitis, with a SCC number greater of 200,000 cells/ml and/or culture positive for udder pathogens, without clinical signs of mastitis. Bacterial DNA was purified and the 16S rRNA genes were individually amplified and sequenced. Significant differences were found in milk samples from healthy quarters and those with sub-clinical and clinical mastitis. The microbiota diversity of milk from healthy quarters was richer as compared to samples with sub-clinical mastitis, whose microbiota diversity was in turn richer as compared to those from clinical mastitis. The core microbiota of water buffalo milk, defined as the asset of microorganisms shared by all healthy milk samples, includes 15 genera, namely Micrococcus, Propionibacterium, 5-7N15, Solibacillus, Staphylococcus, Aerococcus, Facklamia, Trichococcus, Turicibacter, 02d06, SMB53, Clostridium, Acinetobacter, Psychrobacter and Pseudomonas. Only two genera (Acinetobacter and Pseudomonas) were present in all the samples from sub-clinical mastitis, and no genus was shared across all in clinical mastitis milk samples. The presence of mastitis was found to be related to the change in the relative abundance of genera, such as Psychrobacter, whose relative abundance decreased from 16.26% in the milk samples from healthy quarters to 3.2% in clinical mastitis. Other genera, such as SMB53 and Solibacillus, were decreased as well. Discriminant analysis presents the evidence that the microbial community of healthy and clinical mastitis could be discriminated on the background of their microbiota profiles.
Conflict of interest statement
Figures
Similar articles
-
Microbiota of cow's milk; distinguishing healthy, sub-clinically and clinically diseased quarters.PLoS One. 2014 Jan 20;9(1):e85904. doi: 10.1371/journal.pone.0085904. eCollection 2014. PLoS One. 2014. PMID: 24465777 Free PMC article.
-
Impact of intramammary inoculation of inactivated Lactobacillus rhamnosus and antibiotics on the milk microbiota of water buffalo with subclinical mastitis.PLoS One. 2019 Jan 7;14(1):e0210204. doi: 10.1371/journal.pone.0210204. eCollection 2019. PLoS One. 2019. PMID: 30615691 Free PMC article.
-
Microbiological profiles in clinical and subclinical cases of mastitis in milking Jafarabadi buffalo.Res Vet Sci. 2019 Aug;125:94-99. doi: 10.1016/j.rvsc.2019.05.012. Epub 2019 May 22. Res Vet Sci. 2019. PMID: 31176264
-
Review of the bacterial composition of healthy milk, mastitis milk and colostrum in small ruminants.Res Vet Sci. 2021 Nov;140:1-5. doi: 10.1016/j.rvsc.2021.07.022. Epub 2021 Jul 27. Res Vet Sci. 2021. PMID: 34358776 Review.
-
Integrating the milk microbiome signatures in mastitis: milk-omics and functional implications.World J Microbiol Biotechnol. 2025 Jan 18;41(2):41. doi: 10.1007/s11274-024-04242-1. World J Microbiol Biotechnol. 2025. PMID: 39826029 Free PMC article. Review.
Cited by
-
Distinguishing the milk microbiota of healthy goats and goats diagnosed with subclinical mastitis, clinical mastitis, and gangrenous mastitis.Front Microbiol. 2022 Aug 25;13:918706. doi: 10.3389/fmicb.2022.918706. eCollection 2022. Front Microbiol. 2022. PMID: 36090116 Free PMC article.
-
Core microbiome and bacterial diversity of the Italian Mediterranean river buffalo milk.Appl Microbiol Biotechnol. 2023 Mar;107(5-6):1875-1886. doi: 10.1007/s00253-023-12415-5. Epub 2023 Feb 11. Appl Microbiol Biotechnol. 2023. PMID: 36773061
-
A culture-, amplification-independent, and rapid method for identification of pathogens and antibiotic resistance profile in bovine mastitis milk.Front Microbiol. 2023 Jan 6;13:1104701. doi: 10.3389/fmicb.2022.1104701. eCollection 2022. Front Microbiol. 2023. PMID: 36687564 Free PMC article.
-
Peptidomic changes in the milk of water buffaloes (Bubalus bubalis) with intramammary infection by non-aureus staphylococci.Sci Rep. 2022 May 19;12(1):8371. doi: 10.1038/s41598-022-12297-z. Sci Rep. 2022. PMID: 35589845 Free PMC article.
-
The microbiota of Mozzarella di Bufala Campana PDO cheese: a study across the manufacturing process.Front Microbiol. 2023 Aug 15;14:1196879. doi: 10.3389/fmicb.2023.1196879. eCollection 2023. Front Microbiol. 2023. PMID: 37649628 Free PMC article.
References
-
- Stubbendieck RM, Vargas-Bautista C, Straight PD. Bacterial Communities: Interactions to Scale. Front Microbiol. 2016;7: 1234 doi: 10.3389/fmicb.2016.01234 - DOI - PMC - PubMed
-
- Thaiss CA, Zmora N, Levy M, Elinav E. The microbiome and innate immunity. Nature. 2016;535: 65–74. doi: 10.1038/nature18847 - DOI - PubMed
-
- Rogers GB, Hoffman LR, Carroll MP, Bruce KD. Interpreting infective microbiota: the importance of an ecological perspective. Trends Microbiol. 2013;21: 271–276. doi: 10.1016/j.tim.2013.03.004 - DOI - PMC - PubMed
-
- Vayssier-Taussat M, Albina E, Citti C, Cosson JF, Jacques MA, Lebrun MH, et al. Shifting the paradigm from pathogens to pathobiome: new concepts in the light of meta-omics. Front Cell Infect Microbiol. 2014;4: 29 doi: 10.3389/fcimb.2014.00029 - DOI - PMC - PubMed
-
- Weimer PJ. Redundancy, resilience, and host specificity of the ruminal microbiota: implications for engineering improved ruminal fermentations. Front Microbiol. 2015;6: 296 doi: 10.3389/fmicb.2015.00296 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

