The Roles of the SNARE Protein Sed5 in Autophagy in Saccharomyces cerevisiae
- PMID: 28927260
- PMCID: PMC5638772
- DOI: 10.14348/molcells.2017.0030
The Roles of the SNARE Protein Sed5 in Autophagy in Saccharomyces cerevisiae
Abstract
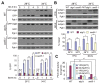
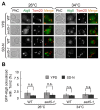
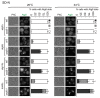
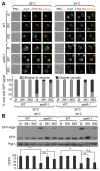
Autophagy is a degradation pathway in eukaryotic cells in which aging proteins and organelles are sequestered into double-membrane vesicles, termed autophagosomes, which fuse with vacuoles to hydrolyze cargo. The key step in autophagy is the formation of autophagosomes, which requires different kinds of vesicles, including COPII vesicles and Atg9-containing vesicles, to transport lipid double-membranes to the phagophore assembly site (PAS). In yeast, the cis-Golgi localized t-SNARE protein Sed5 plays a role in endoplasmic reticulum (ER)-Golgi and intra-Golgi vesicular transport. We report that during autophagy, sed5-1 mutant cells could not properly transport Atg8 to the PAS, resulting in multiple Atg8 dots being dispersed into the cytoplasm. Some dots were trapped in the Golgi apparatus. Sed5 regulates the antero-grade trafficking of Atg9-containing vesicles to the PAS by participating in the localization of Atg23 and Atg27 to the Golgi apparatus. Furthermore, we found that overexpression of SFT1 or SFT2 (suppressor of sed5 ts) rescued the autophagy defects in sed5-1 mutant cells. Our data suggest that Sed5 plays a novel role in autophagy, by regulating the formation of Atg9-containing vesicles in the Golgi apparatus, and the genetic interaction between Sft1/2 and Sed5 is essential for autophagy.
Keywords: Atg9-containing vesicles; Sed5; Sft1/2; autophagy; golgi apparatus.
Figures




Similar articles
-
Autophagy requires Tip20 in Saccharomyces cerevisiae.J Biosci. 2019 Mar;44(1):17. J Biosci. 2019. PMID: 30837368
-
An ER-Localized SNARE Protein Is Exported in Specific COPII Vesicles for Autophagosome Biogenesis.Cell Rep. 2016 Feb 23;14(7):1710-1722. doi: 10.1016/j.celrep.2016.01.047. Epub 2016 Feb 11. Cell Rep. 2016. PMID: 26876173
-
SNARE selectivity of the COPII coat.Cell. 2003 Aug 22;114(4):483-95. doi: 10.1016/s0092-8674(03)00608-1. Cell. 2003. PMID: 12941276
-
The Role of ATG9 Vesicles in Autophagosome Biogenesis.J Mol Biol. 2024 Aug 1;436(15):168489. doi: 10.1016/j.jmb.2024.168489. Epub 2024 Feb 10. J Mol Biol. 2024. PMID: 38342428 Review.
-
Syntaxin 16's Newly Deciphered Roles in Autophagy.Cells. 2019 Dec 17;8(12):1655. doi: 10.3390/cells8121655. Cells. 2019. PMID: 31861136 Free PMC article. Review.
Cited by
-
Atg9-centered multi-omics integration reveals new autophagy regulators in Saccharomyces cerevisiae.Autophagy. 2021 Dec;17(12):4453-4476. doi: 10.1080/15548627.2021.1898749. Epub 2021 Mar 15. Autophagy. 2021. PMID: 33722159 Free PMC article.
-
The Golgin Protein RUD3 Regulates Fusarium graminearum Growth and Virulence.Appl Environ Microbiol. 2021 Feb 26;87(6):e02522-20. doi: 10.1128/AEM.02522-20. Print 2021 Feb 26. Appl Environ Microbiol. 2021. PMID: 33452023 Free PMC article.
-
Autophagy requires Tip20 in Saccharomyces cerevisiae.J Biosci. 2019 Mar;44(1):17. J Biosci. 2019. PMID: 30837368
-
Acetyl-CoA carboxylase 1-dependent lipogenesis promotes autophagy downstream of AMPK.J Biol Chem. 2019 Aug 9;294(32):12020-12039. doi: 10.1074/jbc.RA118.007020. Epub 2019 Jun 17. J Biol Chem. 2019. PMID: 31209110 Free PMC article.
References
-
- Banfield D.K., Lewis M.J., Pelham H.R. A SNARE-like protein required for traffic through the Golgi complex. Nature. 1995;375:806–809. - PubMed
-
- Barlowe C., Orci L., Yeung T., Hosobuchi M., Hamamoto S., Salama N., Rexach M.F., Ravazzola M., Amherdt M., Schekman R. COPII, a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 1994;77:895–907. - PubMed
-
- Cai H., Yu S., Menon S., Cai Y., Lazarova D., Fu C., Reinisch K., Hay J.C., Ferro-Novick S. TRAPPI tethers COPII vesicles by binding the coat subunit Sec23. Nature. 2007;445:941–944. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

