Genome sequencing of the staple food crop white Guinea yam enables the development of a molecular marker for sex determination
- PMID: 28927400
- PMCID: PMC5604175
- DOI: 10.1186/s12915-017-0419-x
Genome sequencing of the staple food crop white Guinea yam enables the development of a molecular marker for sex determination
Abstract
Background: Root and tuber crops are a major food source in tropical Africa. Among these crops are several species in the monocotyledonous genus Dioscorea collectively known as yam, a staple tuber crop that contributes enormously to the subsistence and socio-cultural lives of millions of people, principally in West and Central Africa. Yam cultivation is constrained by several factors, and yam can be considered a neglected "orphan" crop that would benefit from crop improvement efforts. However, the lack of genetic and genomic tools has impeded the improvement of this staple crop.
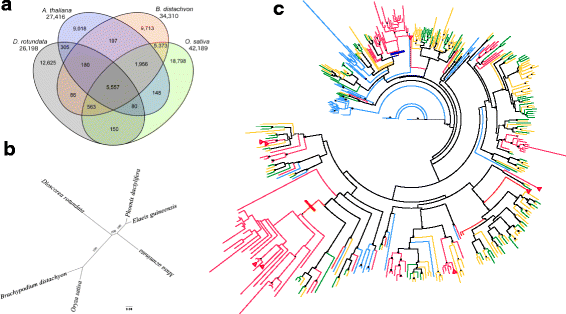
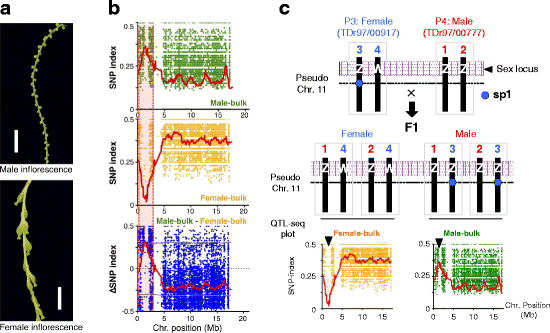
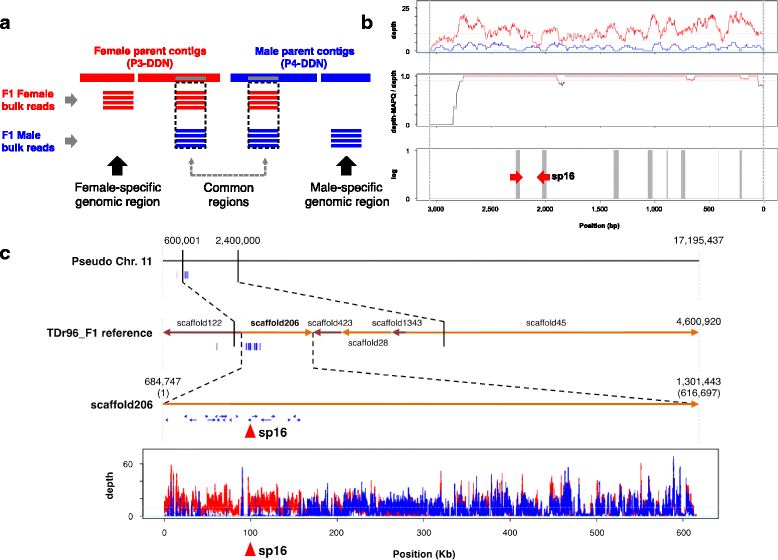
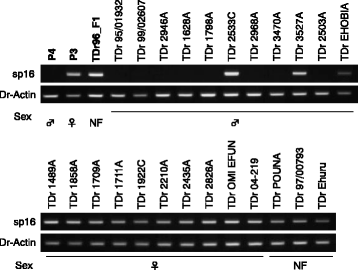
Results: To accelerate marker-assisted breeding of yam, we performed genome analysis of white Guinea yam (Dioscorea rotundata) and assembled a 594-Mb genome, 76.4% of which was distributed among 21 linkage groups. In total, we predicted 26,198 genes. Phylogenetic analyses with 2381 conserved genes revealed that Dioscorea is a unique lineage of monocotyledons distinct from the Poales (rice), Arecales (palm), and Zingiberales (banana). The entire Dioscorea genus is characterized by the occurrence of separate male and female plants (dioecy), a feature that has limited efficient yam breeding. To infer the genetics of sex determination, we performed whole-genome resequencing of bulked segregants (quantitative trait locus sequencing [QTL-seq]) in F1 progeny segregating for male and female plants and identified a genomic region associated with female heterogametic (male = ZZ, female = ZW) sex determination. We further delineated the W locus and used it to develop a molecular marker for sex identification of Guinea yam plants at the seedling stage.
Conclusions: Guinea yam belongs to a unique and highly differentiated clade of monocotyledons. The genome analyses and sex-linked marker development performed in this study should greatly accelerate marker-assisted breeding of Guinea yam. In addition, our QTL-seq approach can be utilized in genetic studies of other outcrossing crops and organisms with highly heterozygous genomes. Genomic analysis of orphan crops such as yam promotes efforts to improve food security and the sustainability of tropical agriculture.
Keywords: Dioecy; Dioscorea; Sex determination; Whole-genome sequence; Yam.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures




References
-
- Wilkin P, Scholsb P, Chasea MW, Chayamaritc K, Furnessa CA, Huysmansb S, Rakotonasolod F, et al. A plastid gene phylogeny of the yam genus, Dioscorea: roots, fruits and Madagascar. Syst Bot. 2005;30:736–49. doi: 10.1600/036364405775097879. - DOI
-
- Lebot V. Tropical root and tuber crops: cassava, sweet potato, yams and aroids (Crop Production Science in Horticulture Series 17) Wallingford: CABI Publishing; 2009. p. 405.
-
- Coursey DG. The civilizations of the yam: interrelationships of man and yams in Africa and the Indo-Pacific region. Archeol Phys Anthropol Oceania. 1972;7:215–33.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

