First molecular detection and characterization of zoonotic Bartonella species in fleas infesting domestic animals in Tunisia
- PMID: 28927427
- PMCID: PMC5606017
- DOI: 10.1186/s13071-017-2372-5
First molecular detection and characterization of zoonotic Bartonella species in fleas infesting domestic animals in Tunisia
Abstract
Background: Bartonellosis is an emerging vector-borne disease caused by different intracellular bacteria of the genus Bartonella (Rhizobiales: Bartonellaceae) that is transmitted primarily by blood-sucking arthropods such as sandflies, ticks and fleas. In Tunisia, there are no data available identifying the vectors of Bartonella spp. In our research, we used molecular methods to detect and characterize Bartonella species circulating in fleas collected from domestic animals in several of the country's bioclimatic areas.
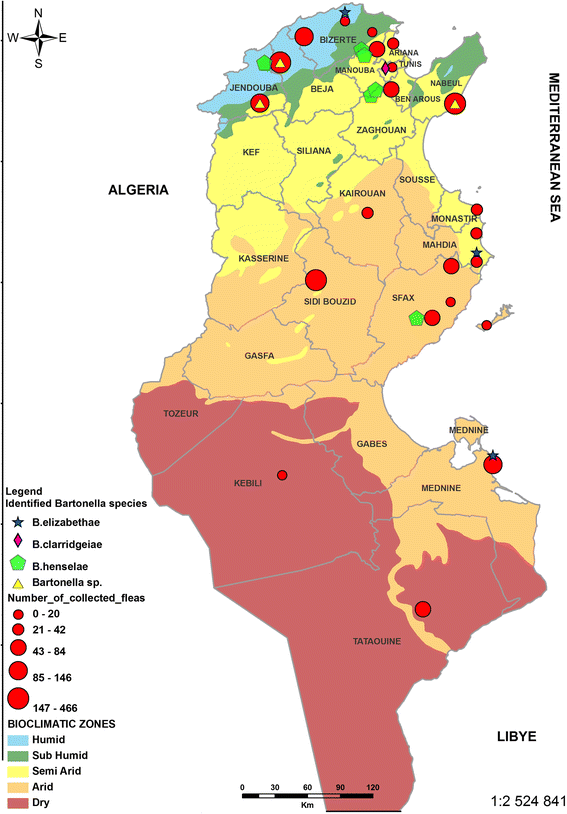
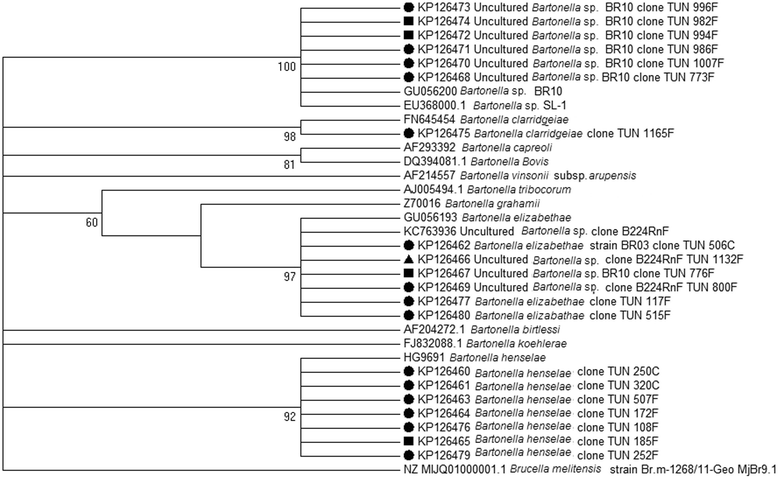
Results: A total of 2178 fleas were collected from 5 cats, 27 dogs, 34 sheep, and 41 goats at 22 sites located in Tunisia's five bioclimatic zones. The fleas were identified as: 1803 Ctenocephalides felis (83%) (Siphonaptera: Pulicidae), 266 C. canis (12%) and 109 Pulex irritans (5%) (Siphonaptera: Pulicidae). Using conventional PCR, we screened the fleas for the presence of Bartonella spp., targeting the citrate synthase gene (gltA). Bartonella DNA was detected in 14% (121/866) of the tested flea pools [estimated infection rate (EIR) per 2 specimens: 0.072, 95% confidence interval (CI): 0.060-0.086]. The Bartonella infection rate per pool was broken down as follows: 55% (65/118; EIR per 2 specimens: 0.329, 95% CI: 0.262-0.402) in C. canis; 23.5% (8/34; EIR per 2 specimens: 0.125, 95% CI: 0.055-0.233) in P. irritans and 6.7% (48/714; EIR per 2 specimens: 0.032, 95% CI: 0.025-0.045) in C. felis. Infection rates, which varied significantly by bioclimatic zone (P < 0.0001), were highest in the humid areas. By sequencing, targeting the gltA gene and the 16S-23S rRNA Intergenic Spacer Regions (ITS), we identified three Bartonella zoonotic species: B. elizabethae, B. henselae, B. clarridgeiae, as well as uncharacterized Bartonella genotypes.
Conclusions: To the best of our knowledge, this is the first time that fleas in Tunisia have been shown to carry zoonotic species of Bartonella. The dog flea, Ctenocephalides canis, should be considered the main potential vector of Bartonella. Our study not only provides new information about this vector, but also offers a public health update: medical practitioners and farmers in Tunisia should be apprised of the presence of Bartonella in fleas and implement preventive measures.
Keywords: B. clarridgeiae; B. henselae; Bartonella elizabethae; Bartonellosis; C. canis; Ctenocephalides felis; Pulex irritans; Tunisia; Zoonotic Bartonella species.
Conflict of interest statement
Ethics approval and consent to participate
This study is a part of the “PROJET 2 PS1.3.023-RESTUS” and has been approved by the Commission on Ethics and Animal Welfare of the Institut Pasteur de Tunis - Université de Tunis El Manar-TUNISIA, number 2014/03/I/LR11IPT03/V1. All technical procedures were in accordance with the National and the European legislation regarding animal welfare and have met the International Guiding Principles for Biomedical Research Involving Animals by the Council for the International Organizations of Medical Sciences.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
Molecular detection of zoonotic bartonellae (B. henselae, B. elizabethae and B. rochalimae) in fleas collected from dogs in Israel.Med Vet Entomol. 2015 Sep;29(3):344-8. doi: 10.1111/mve.12116. Epub 2015 Apr 11. Med Vet Entomol. 2015. PMID: 25865162
-
Molecular Detection of Bartonella Species in Fleas Collected from Dogs and Cats from Costa Rica.Vector Borne Zoonotic Dis. 2015 Oct;15(10):630-2. doi: 10.1089/vbz.2015.1799. Epub 2015 Sep 22. Vector Borne Zoonotic Dis. 2015. PMID: 26393956
-
Flea species infesting dogs in Florida and Bartonella spp. prevalence rates.Vet Parasitol. 2014 Jan 31;199(3-4):225-9. doi: 10.1016/j.vetpar.2013.10.017. Epub 2013 Oct 30. Vet Parasitol. 2014. PMID: 24268654
-
A case of mistaken identity: a systematic review, meta-analysis, and reinvestigation of hemotropic Mycoplasma spp. infection in Ctenocephalides felis (cat flea).Parasit Vectors. 2024 May 9;17(1):209. doi: 10.1186/s13071-024-06292-8. Parasit Vectors. 2024. PMID: 38720359 Free PMC article.
-
Bartonella spp. in pets and effect on human health.Emerg Infect Dis. 2006 Mar;12(3):389-94. doi: 10.3201/eid1203.050931. Emerg Infect Dis. 2006. PMID: 16704774 Free PMC article. Review.
Cited by
-
Detection of Bartonella henselae DNA in Triatoma sordida collected in peridomiciliary environments.Braz J Infect Dis. 2024 Sep-Oct;28(5):103875. doi: 10.1016/j.bjid.2024.103875. Epub 2024 Sep 28. Braz J Infect Dis. 2024. PMID: 39353573 Free PMC article.
-
Flea surveillance on wild mammals in northern region of Xinjiang, northwestern China.Int J Parasitol Parasites Wildl. 2019 Dec 1;11:12-16. doi: 10.1016/j.ijppaw.2019.11.004. eCollection 2020 Apr. Int J Parasitol Parasites Wildl. 2019. PMID: 31879590 Free PMC article.
-
Molecular Evidence of a Broad Range of Pathogenic Bacteria in Ctenocephalides spp.: Should We Re-Examine the Role of Fleas in the Transmission of Pathogens?Trop Med Infect Dis. 2021 Mar 17;6(1):37. doi: 10.3390/tropicalmed6010037. Trop Med Infect Dis. 2021. PMID: 33802921 Free PMC article.
-
Bartonella Infections in Cats and Cat Fleas in Lithuania.Pathogens. 2021 Sep 17;10(9):1209. doi: 10.3390/pathogens10091209. Pathogens. 2021. PMID: 34578241 Free PMC article.
-
Molecular Evidence of Bartonella melophagi in Ticks in Border Areas of Xinjiang, China.Front Vet Sci. 2021 Jun 22;8:675457. doi: 10.3389/fvets.2021.675457. eCollection 2021. Front Vet Sci. 2021. PMID: 34239911 Free PMC article.
References
-
- Boulouis PH-J, Marignac G, Haddad N, Maillard R, Chomel B. Animal reservoirs and primary hosts of Bartonella. Bull Acad Veterinaire Fr. 2008;161:211–220. doi: 10.4267/2042/47945. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

