SecA mediates cotranslational targeting and translocation of an inner membrane protein
- PMID: 28928132
- PMCID: PMC5674894
- DOI: 10.1083/jcb.201704036
SecA mediates cotranslational targeting and translocation of an inner membrane protein
Abstract
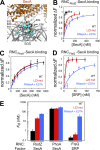
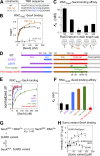
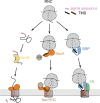
Protein targeting to the bacterial plasma membrane was generally thought to occur via two major pathways: cotranslational targeting by signal recognition particle (SRP) and posttranslational targeting by SecA and SecB. Recently, SecA was found to also bind ribosomes near the nascent polypeptide exit tunnel, but the function of this SecA-ribosome contact remains unclear. In this study, we show that SecA cotranslationally recognizes the nascent chain of an inner membrane protein, RodZ, with high affinity and specificity. In vitro reconstitution and in vivo targeting assays show that SecA is necessary and sufficient to direct the targeting and translocation of RodZ to the bacterial plasma membrane in an obligatorily cotranslational mechanism. Sequence elements upstream and downstream of the RodZ transmembrane domain dictate nascent polypeptide selection by SecA instead of the SRP machinery. These findings identify a new route for the targeting of inner membrane proteins in bacteria and highlight the diversity of targeting pathways that enables an organism to accommodate diverse nascent proteins.
© 2017 Wang et al.
Figures




References
-
- Akita M., Sasaki S., Matsuyama S., and Mizushima S.. 1990. SecA interacts with secretory proteins by recognizing the positive charge at the amino terminus of the signal peptide in Escherichia coli. J. Biol. Chem. 265:8164–8169. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

