The CRISPR Spacer Space Is Dominated by Sequences from Species-Specific Mobilomes
- PMID: 28928211
- PMCID: PMC5605939
- DOI: 10.1128/mBio.01397-17
The CRISPR Spacer Space Is Dominated by Sequences from Species-Specific Mobilomes
Abstract
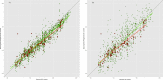
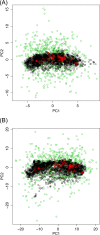
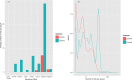
Clustered regularly interspaced short palindromic repeats and CRISPR-associated protein (CRISPR-Cas) systems store the memory of past encounters with foreign DNA in unique spacers that are inserted between direct repeats in CRISPR arrays. For only a small fraction of the spacers, homologous sequences, called protospacers, are detectable in viral, plasmid, and microbial genomes. The rest of the spacers remain the CRISPR "dark matter." We performed a comprehensive analysis of the spacers from all CRISPR-cas loci identified in bacterial and archaeal genomes, and we found that, depending on the CRISPR-Cas subtype and the prokaryotic phylum, protospacers were detectable for 1% to about 19% of the spacers (~7% global average). Among the detected protospacers, the majority, typically 80 to 90%, originated from viral genomes, including proviruses, and among the rest, the most common source was genes that are integrated into microbial chromosomes but are involved in plasmid conjugation or replication. Thus, almost all spacers with identifiable protospacers target mobile genetic elements (MGE). The GC content, as well as dinucleotide and tetranucleotide compositions, of microbial genomes, their spacer complements, and the cognate viral genomes showed a nearly perfect correlation and were almost identical. Given the near absence of self-targeting spacers, these findings are most compatible with the possibility that the spacers, including the dark matter, are derived almost completely from the species-specific microbial mobilomes.IMPORTANCE The principal function of CRISPR-Cas systems is thought to be protection of bacteria and archaea against viruses and other parasitic genetic elements. The CRISPR defense function is mediated by sequences from parasitic elements, known as spacers, that are inserted into CRISPR arrays and then transcribed and employed as guides to identify and inactivate the cognate parasitic genomes. However, only a small fraction of the CRISPR spacers match any sequences in the current databases, and of these, only a minority correspond to known parasitic elements. We show that nearly all spacers with matches originate from viral or plasmid genomes that are either free or have been integrated into the host genome. We further demonstrate that spacers with no matches have the same properties as those of identifiable origins, strongly suggesting that all spacers originate from mobile elements.
Keywords: CRISPR-Cas; bacteriophages; mobilome; oligonucleotide composition; spacer acquisition.
Figures
Similar articles
-
CRISPRCasdb a successor of CRISPRdb containing CRISPR arrays and cas genes from complete genome sequences, and tools to download and query lists of repeats and spacers.Nucleic Acids Res. 2020 Jan 8;48(D1):D535-D544. doi: 10.1093/nar/gkz915. Nucleic Acids Res. 2020. PMID: 31624845 Free PMC article.
-
On the Origin of Reverse Transcriptase-Using CRISPR-Cas Systems and Their Hyperdiverse, Enigmatic Spacer Repertoires.mBio. 2017 Jul 11;8(4):e00897-17. doi: 10.1128/mBio.00897-17. mBio. 2017. PMID: 28698278 Free PMC article.
-
Survey of clustered regularly interspaced short palindromic repeats and their associated Cas proteins (CRISPR/Cas) systems in multiple sequenced strains of Klebsiella pneumoniae.BMC Res Notes. 2015 Aug 4;8:332. doi: 10.1186/s13104-015-1285-7. BMC Res Notes. 2015. PMID: 26238567 Free PMC article.
-
Mobile Genetic Elements and Evolution of CRISPR-Cas Systems: All the Way There and Back.Genome Biol Evol. 2017 Oct 1;9(10):2812-2825. doi: 10.1093/gbe/evx192. Genome Biol Evol. 2017. PMID: 28985291 Free PMC article. Review.
-
Molecular mechanisms of CRISPR-Cas spacer acquisition.Nat Rev Microbiol. 2019 Jan;17(1):7-12. doi: 10.1038/s41579-018-0071-7. Nat Rev Microbiol. 2019. PMID: 30171202 Review.
Cited by
-
Identification of an anti-CRISPR protein that inhibits the CRISPR-Cas type I-B system in Clostridioides difficile.mSphere. 2023 Dec 20;8(6):e0040123. doi: 10.1128/msphere.00401-23. Epub 2023 Nov 27. mSphere. 2023. PMID: 38009936 Free PMC article.
-
Dynamics of immune memory and learning in bacterial communities.Elife. 2023 Jan 16;12:e81692. doi: 10.7554/eLife.81692. Elife. 2023. PMID: 36645771 Free PMC article.
-
Global phylogenomic novelty of the Cas1 gene from hot spring microbial communities.Front Microbiol. 2022 Dec 2;13:1069452. doi: 10.3389/fmicb.2022.1069452. eCollection 2022. Front Microbiol. 2022. PMID: 36532491 Free PMC article.
-
Genomic diversity of genus Limosilactobacillus.Microb Genom. 2022 Jul;8(7):mgen000847. doi: 10.1099/mgen.0.000847. Microb Genom. 2022. PMID: 35838756 Free PMC article.
-
The CRISPR-Cas systems were selectively inactivated during evolution of Bacillus cereus group for adaptation to diverse environments.ISME J. 2020 Jun;14(6):1479-1493. doi: 10.1038/s41396-020-0623-5. Epub 2020 Mar 4. ISME J. 2020. PMID: 32132663 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

