The Ubiquitin Ligase (E3) Psh1p Is Required for Proper Segregation of both Centromeric and Two-Micron Plasmids in Saccharomyces cerevisiae
- PMID: 28928274
- PMCID: PMC5677152
- DOI: 10.1534/g3.117.300227
The Ubiquitin Ligase (E3) Psh1p Is Required for Proper Segregation of both Centromeric and Two-Micron Plasmids in Saccharomyces cerevisiae
Abstract
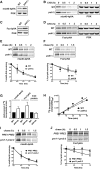
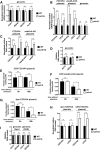
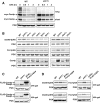
Protein degradation by the ubiquitin-proteasome system is essential to many processes. We sought to assess its involvement in the turnover of mitochondrial proteins in Saccharomyces cerevisiae We find that deletion of a specific ubiquitin ligase (E3), Psh1p, increases the abundance of a temperature-sensitive mitochondrial protein, mia40-4pHA, when it is expressed from a centromeric plasmid. Deletion of Psh1p unexpectedly elevates the levels of other proteins expressed from centromeric plasmids. Loss of Psh1p does not increase the rate of turnover of mia40-4pHA, affect total protein synthesis, or increase the protein levels of chromosomal genes. Instead, psh1Δ appears to increase the incidence of missegregation of centromeric plasmids relative to their normal 1:1 segregation. After generations of growth with selection for the plasmid, ongoing missegregation would lead to elevated plasmid DNA, mRNA, and protein, all of which we observe in psh1Δ cells. The only known substrate of Psh1p is the centromeric histone H3 variant Cse4p, which is targeted for proteasomal degradation after ubiquitination by Psh1p However, Cse4p overexpression alone does not phenocopy psh1Δ in increasing plasmid DNA and protein levels. Instead, elevation of Cse4p leads to an apparent increase in 1:0 plasmid segregation events. Further, 2 μm high-copy yeast plasmids also missegregate in psh1Δ, but not when Cse4p alone is overexpressed. These findings demonstrate that Psh1p is required for the faithful inheritance of both centromeric and 2 μm plasmids. Moreover, the effects that loss of Psh1p has on plasmid segregation cannot be accounted for by increased levels of Cse4p.
Keywords: 2 μm plasmid; CEN plasmid; Cse4p; plasmid missegregation; ubiquitination.
Copyright © 2017 Metzger et al.
Figures




Similar articles
-
The centromere-specific histone variant Cse4p (CENP-A) is essential for functional chromatin architecture at the yeast 2-microm circle partitioning locus and promotes equal plasmid segregation.J Cell Biol. 2006 Sep 11;174(6):779-90. doi: 10.1083/jcb.200603042. J Cell Biol. 2006. PMID: 16966420 Free PMC article.
-
Molecular basis for the selective recognition and ubiquitination of centromeric histone H3 by yeast E3 ligase Psh1.J Genet Genomics. 2021 Jun 20;48(6):463-472. doi: 10.1016/j.jgg.2021.04.007. Epub 2021 May 26. J Genet Genomics. 2021. PMID: 34217622
-
Cse4 gets a kiss-of-death from Psh1.Cell Cycle. 2011 Feb 15;10(4):566-7. doi: 10.4161/cc.10.4.14770. Epub 2011 Feb 15. Cell Cycle. 2011. PMID: 21293184 No abstract available.
-
A hitchhiker's guide to survival finally makes CENs.J Cell Biol. 2006 Sep 11;174(6):747-9. doi: 10.1083/jcb.200608107. J Cell Biol. 2006. PMID: 16966417 Free PMC article. Review.
-
The 2 micron plasmid: a selfish genetic element with an optimized survival strategy within Saccharomyces cerevisiae.Curr Genet. 2018 Feb;64(1):25-42. doi: 10.1007/s00294-017-0719-2. Epub 2017 Jun 8. Curr Genet. 2018. PMID: 28597305 Review.
Cited by
-
Suppression of chromosome instability by targeting a DNA helicase in budding yeast.Mol Biol Cell. 2023 Jan 1;34(1):ar3. doi: 10.1091/mbc.E22-09-0395. Epub 2022 Nov 9. Mol Biol Cell. 2023. PMID: 36350688 Free PMC article.
-
Chromatin assembly factor-1 (CAF-1) chaperone regulates Cse4 deposition into chromatin in budding yeast.Nucleic Acids Res. 2018 May 18;46(9):4440-4455. doi: 10.1093/nar/gky169. Nucleic Acids Res. 2018. PMID: 29522205 Free PMC article.
-
Gene Amplification as a Mechanism of Yeast Adaptation to Nonsense Mutations in Release Factor Genes.Genes (Basel). 2021 Dec 19;12(12):2019. doi: 10.3390/genes12122019. Genes (Basel). 2021. PMID: 34946968 Free PMC article.
-
Formyl-methionine as an N-degron of a eukaryotic N-end rule pathway.Science. 2018 Nov 30;362(6418):eaat0174. doi: 10.1126/science.aat0174. Epub 2018 Nov 8. Science. 2018. PMID: 30409808 Free PMC article.
-
A Genome-Wide Screen Reveals a Role for the HIR Histone Chaperone Complex in Preventing Mislocalization of Budding Yeast CENP-A.Genetics. 2018 Sep;210(1):203-218. doi: 10.1534/genetics.118.301305. Epub 2018 Jul 16. Genetics. 2018. PMID: 30012561 Free PMC article.
References
-
- Akutsu M., Dikic I., Bremm A., 2016. Ubiquitin chain diversity at a glance. J. Cell Sci. 129: 875–880. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
