Tumor-associated B-cells induce tumor heterogeneity and therapy resistance
- PMID: 28928360
- PMCID: PMC5605714
- DOI: 10.1038/s41467-017-00452-4
Tumor-associated B-cells induce tumor heterogeneity and therapy resistance
Abstract
In melanoma, therapies with inhibitors to oncogenic BRAFV600E are highly effective but responses are often short-lived due to the emergence of drug-resistant tumor subpopulations. We describe here a mechanism of acquired drug resistance through the tumor microenvironment, which is mediated by human tumor-associated B cells. Human melanoma cells constitutively produce the growth factor FGF-2, which activates tumor-infiltrating B cells to produce the growth factor IGF-1. B-cell-derived IGF-1 is critical for resistance of melanomas to BRAF and MEK inhibitors due to emergence of heterogeneous subpopulations and activation of FGFR-3. Consistently, resistance of melanomas to BRAF and/or MEK inhibitors is associated with increased CD20 and IGF-1 transcript levels in tumors and IGF-1 expression in tumor-associated B cells. Furthermore, first clinical data from a pilot trial in therapy-resistant metastatic melanoma patients show anti-tumor activity through B-cell depletion by anti-CD20 antibody. Our findings establish a mechanism of acquired therapy resistance through tumor-associated B cells with important clinical implications.Resistance to BRAFV600E inhibitors often occurs in melanoma patients. Here, the authors describe a potential mechanism of acquired drug resistance mediated by tumor-associated B cells-derived IGF-1.
Conflict of interest statement
G.L. has consulting or advisory role for GSK, BMS, Novartis, Genentech, Amgen, Merck, Pierre Fabre, Array, and Roche. K.F. has consulting or advisory role for Novartis, Roche, and Array. J.W. is an advisory board member for GSK, Roche/Genentech, Novartis, and BMS. M.A.D. is an advisory board member for GSK, Roche/Genentech, Novartis, Sanofi, and Vaccinex. R.Sc. received an honorarium from Roche. The remaining authors declare no competing financial interests.
Figures
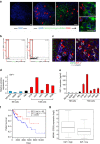
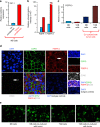



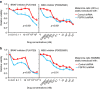

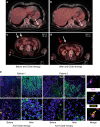
References
-
- Schadendorf D, et al. Melanoma. Nat. Rev. Dis. Primers. 2015;1:1–20. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

