RecA-SSB Interaction Modulates RecA Nucleoprotein Filament Formation on SSB-Wrapped DNA
- PMID: 28928411
- PMCID: PMC5605508
- DOI: 10.1038/s41598-017-12213-w
RecA-SSB Interaction Modulates RecA Nucleoprotein Filament Formation on SSB-Wrapped DNA
Abstract
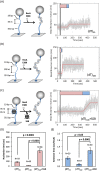
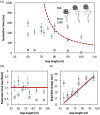
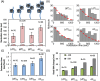
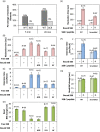
E. coli RecA recombinase catalyzes the homology pairing and strand exchange reactions in homologous recombinational repair. RecA must compete with single-stranded DNA binding proteins (SSB) for single-stranded DNA (ssDNA) substrates to form RecA nucleoprotein filaments, as the first step of this repair process. It has been suggested that RecA filaments assemble mainly by binding and extending onto the free ssDNA region not covered by SSB, or are assisted by mediators. Using the tethered particle motion (TPM) technique, we monitored individual RecA filament assembly on SSB-wrapped ssDNA in real-time. Nucleation times of the RecA E38K nucleoprotein filament assembly showed no apparent dependence among DNA substrates with various ssDNA gap lengths (from 60 to 100 nucleotides) wrapped by one SSB in the (SSB)65 binding mode. Our data have shown an unexpected RecA filament assembly mechanism in which a RecA-SSB-ssDNA interaction exists. Four additional pieces of evidence support our claim: the nucleation times of the RecA assembly varied (1) when DNA substrates contained different numbers of bound SSB tetramers; (2) when the SSB wrapping mode conversion is induced; (3) when SSB C-terminus truncation mutants are used; and (4) when an excess of C-terminal peptide of SSB is present. Thus, a RecA-SSB interaction should be included in discussing RecA regulatory mechanism.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
Direct imaging of RecA nucleation and growth on single molecules of SSB-coated ssDNA.Nature. 2012 Nov 8;491(7423):274-8. doi: 10.1038/nature11598. Epub 2012 Oct 24. Nature. 2012. PMID: 23103864 Free PMC article.
-
Stable Nuclei of Nucleoprotein Filament and High ssDNA Binding Affinity Contribute to Enhanced RecA E38K Recombinase Activity.Sci Rep. 2017 Nov 2;7(1):14964. doi: 10.1038/s41598-017-15088-z. Sci Rep. 2017. PMID: 29097773 Free PMC article.
-
The C terminus of the Escherichia coli RecA protein modulates the DNA binding competition with single-stranded DNA-binding protein.J Biol Chem. 2003 May 2;278(18):16389-96. doi: 10.1074/jbc.M212920200. Epub 2003 Feb 20. J Biol Chem. 2003. PMID: 12598538
-
RecA and DNA recombination: a review of molecular mechanisms.Biochem Soc Trans. 2019 Oct 31;47(5):1511-1531. doi: 10.1042/BST20190558. Biochem Soc Trans. 2019. PMID: 31654073 Review.
-
RecA: Regulation and Mechanism of a Molecular Search Engine.Trends Biochem Sci. 2016 Jun;41(6):491-507. doi: 10.1016/j.tibs.2016.04.002. Epub 2016 May 4. Trends Biochem Sci. 2016. PMID: 27156117 Free PMC article. Review.
Cited by
-
Structural Adaptation of the Single-Stranded DNA-Binding Protein C-Terminal to DNA Metabolizing Partners Guides Inhibitor Design.Pharmaceutics. 2023 Mar 23;15(4):1032. doi: 10.3390/pharmaceutics15041032. Pharmaceutics. 2023. PMID: 37111518 Free PMC article.
-
SSB Facilitates Fork-Substrate Discrimination by the PriA DNA Helicase.ACS Omega. 2021 Jun 15;6(25):16324-16335. doi: 10.1021/acsomega.1c00722. eCollection 2021 Jun 29. ACS Omega. 2021. PMID: 34235303 Free PMC article.
-
Activation and modulation of the host response to DNA damage by an integrative and conjugative element.J Bacteriol. 2025 Feb 20;207(2):e0046224. doi: 10.1128/jb.00462-24. Epub 2025 Jan 23. J Bacteriol. 2025. PMID: 39846752 Free PMC article.
-
Microcephaly family protein MCPH1 stabilizes RAD51 filaments.Nucleic Acids Res. 2020 Sep 18;48(16):9135-9146. doi: 10.1093/nar/gkaa636. Nucleic Acids Res. 2020. PMID: 32735676 Free PMC article.
-
The mechanism of action of the SSB interactome reveals it is the first OB-fold family of genome guardians in prokaryotes.Protein Sci. 2021 Sep;30(9):1757-1775. doi: 10.1002/pro.4140. Epub 2021 Jun 14. Protein Sci. 2021. PMID: 34089559 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

