Bacterial communities 16S rDNA fingerprinting as a potential tracing tool for cultured seabass Dicentrarchus labrax
- PMID: 28928412
- PMCID: PMC5605529
- DOI: 10.1038/s41598-017-11552-y
Bacterial communities 16S rDNA fingerprinting as a potential tracing tool for cultured seabass Dicentrarchus labrax
Abstract
Traceability of seafood has become crucial with market globalization and consumer's awareness. The present study used PCR-DGGE and 454 pyrosequencing to assess if bacterial communities fingerprint associated to seabass (Dicentrarchus labrax) skin mucus can be used to discriminate the geographic origin of fishes cultured in three semi-intensive fish farms. PCR-DGGE and pyrosequencing results were congruent and suggested that this molecular approach has the potential to trace fish farms with a spatial resolution <500 m. Pyrosequencing results provided a detailed insight into the bacterial community composition of seabass skin mucus and revealed the existence of a core of bacterial communities within family Pseudomonadaceae and Rhodobacteraceae. This approach also allowed to recognized key OTUs that are potentially relevant to discriminate the geographic origin of the fish being surveyed. Overall, the present study increased our knowledge on farmed seabass microbiome and demonstrated that specific and unique bacterial taxa can act as natural signatures that allow us to trace fish to its respective geographic origin. Our study provides valuable clues that should be more investigated in future studies as a way to fulfill current traceability needs in the global trade of seafood.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

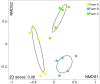
 - Farm A,
- Farm A,  - Farm B and
- Farm B and  - Farm C. The map was created using the software ArcGIS v10.2.2.
- Farm C. The map was created using the software ArcGIS v10.2.2.




Similar articles
-
Detection and identification of a pathogenic marine Mycobacterium from the European seabass Dicentrarchus labrax using polymerase chain reaction and direct sequencing of 16S rDNA sequences.Mol Mar Biol Biotechnol. 1993 Aug;2(4):225-32. Mol Mar Biol Biotechnol. 1993. PMID: 8293073
-
Monitoring Infection and Antibiotic Treatment in the Skin Microbiota of Farmed European Seabass (Dicentrarchus Labrax) Fingerlings.Microb Ecol. 2022 Apr;83(3):789-797. doi: 10.1007/s00248-021-01795-8. Epub 2021 Jul 10. Microb Ecol. 2022. PMID: 34245329
-
Pyrosequencing survey of intestinal microbiota diversity in cultured sea bass (Dicentrarchus labrax) fed functional diets.FEMS Microbiol Ecol. 2014 Feb;87(2):451-9. doi: 10.1111/1574-6941.12236. Epub 2013 Nov 4. FEMS Microbiol Ecol. 2014. PMID: 24730648
-
How to Determine the Geographical Origin of Seafood?Crit Rev Food Sci Nutr. 2016;56(2):306-17. doi: 10.1080/10408398.2012.745478. Crit Rev Food Sci Nutr. 2016. PMID: 24834835 Review.
-
Microbiome Sequencing Methods for Studying Human Diseases.Methods Mol Biol. 2018;1706:77-90. doi: 10.1007/978-1-4939-7471-9_5. Methods Mol Biol. 2018. PMID: 29423794 Review.
Cited by
-
Detection of an amplification bias associated to Leuconostocaceae family with a universal primer routinely used for monitoring microbial community structures within food products.BMC Res Notes. 2018 Nov 8;11(1):802. doi: 10.1186/s13104-018-3908-2. BMC Res Notes. 2018. PMID: 30409220 Free PMC article.
-
Gut microbiota fingerprinting as a potential tool for tracing the geographical origin of farmed mussels (Mytilus galloprovincialis).PLoS One. 2023 Aug 30;18(8):e0290776. doi: 10.1371/journal.pone.0290776. eCollection 2023. PLoS One. 2023. PMID: 37647359 Free PMC article.
-
Improved PCR-DGGE analysis by emulsion-PCR for the determination of food geographical origin: A case study on Greek PDO "avgotaracho Mesolonghiou".Curr Res Food Sci. 2021 Oct 20;4:746-751. doi: 10.1016/j.crfs.2021.10.005. eCollection 2021. Curr Res Food Sci. 2021. PMID: 34746808 Free PMC article.
-
Influences of claywater and greenwater on the skin microbiome of cultured larval sablefish (Anoplopoma fimbria).Anim Microbiome. 2020 Aug 4;2(1):27. doi: 10.1186/s42523-020-00045-5. Anim Microbiome. 2020. PMID: 33499990 Free PMC article.
-
Spatial Fingerprinting: Horizontal Fusion of Multi-Dimensional Bio-Tracers as Solution to Global Food Provenance Problems.Foods. 2021 Mar 28;10(4):717. doi: 10.3390/foods10040717. Foods. 2021. PMID: 33800611 Free PMC article.
References
-
- FAO. Fishery and Aquaculture Statistics. Global aquaculture production 1950–2014 (FishstatJ). In: FAO Fisheries and Aquaculture Department [online or CD-ROM]. Rome. Updated 2017 http://www.fao.org/fishery/statistics/software/fishstatj/en (2016).
-
- El Sheikha AF, Xu J. Traceability as a key of seafood safety: reassessment and possible applications. Rev. Fish. Sci. Aquac. 2017;25:158–170. doi: 10.1080/23308249.2016.1254158. - DOI
-
- Charlebois S, Schwab A, Henn R, Huck CW. Food fraud: An exploratory study for measuring consumer perception towards mislabeled food products and influence on self-authentication intentions. Trends Food Sci. Technol. 2016;50:211–218. doi: 10.1016/j.tifs.2016.02.003. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

