A comprehensive characterization of PncA polymorphisms that confer resistance to pyrazinamide
- PMID: 28928454
- PMCID: PMC5605632
- DOI: 10.1038/s41467-017-00721-2
A comprehensive characterization of PncA polymorphisms that confer resistance to pyrazinamide
Abstract
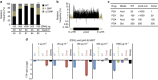
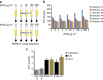
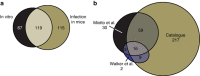
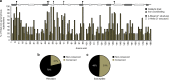
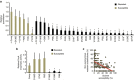
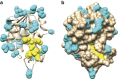
Tuberculosis chemotherapy is dependent on the use of the antibiotic pyrazinamide, which is being threatened by emerging drug resistance. Resistance is mediated through mutations in the bacterial gene pncA. Methods for testing pyrazinamide susceptibility are difficult and rarely performed, and this means that the full spectrum of pncA alleles that confer clinical resistance to pyrazinamide is unknown. Here, we performed in vitro saturating mutagenesis of pncA to generate a comprehensive library of PncA polymorphisms resultant from a single-nucleotide polymorphism. We then screened it for pyrazinamide resistance both in vitro and in an infected animal model. We identify over 300 resistance-conferring substitutions. Strikingly, these mutations map throughout the PncA structure and result in either loss of enzymatic activity and/or decrease in protein abundance. Our comprehensive mutational and screening approach should stand as a paradigm for determining resistance mutations and their mechanisms of action.The antibiotic pyrazinamide is central to tuberculosis treatment regimens, globally. Despite its efficacy, resistance to the drug is increasing. Here, Eric Rubin and colleagues characterise the genetic basis of pyrazinamide resistance.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
References
-
- World Health Organization. Global Tuberculosis Report 2016. Report No. WHO/HTM/TB/2016.13 (World Health Organization, Geneva, 2016).
-
- World Health Organization. Treatment of Tuberculosis Guidlines. Report No. WHO/HTM/TB/2009.420 (World Health Organization, Geneva, 2010).
-
- Dawson R, et al. Efficiency and safety of the combination of moxifloxacin, pretomanid (PA-824), and pyrazinamide during the first 8 weeks of antituberculosis treatment: a phase 2b, open-label, partly randomised trial in patients with drug-susceptible or drug-resistant pulmonary tuberculosis. Lancet. 2015;385:1738–1747. doi: 10.1016/S0140-6736(14)62002-X. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

