Revisiting inconsistency in large pharmacogenomic studies
- PMID: 28928933
- PMCID: PMC5580432
- DOI: 10.12688/f1000research.9611.3
Revisiting inconsistency in large pharmacogenomic studies
Abstract
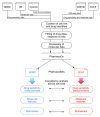
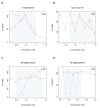
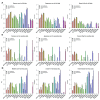
In 2013, we published a comparative analysis of mutation and gene expression profiles and drug sensitivity measurements for 15 drugs characterized in the 471 cancer cell lines screened in the Genomics of Drug Sensitivity in Cancer (GDSC) and Cancer Cell Line Encyclopedia (CCLE). While we found good concordance in gene expression profiles, there was substantial inconsistency in the drug responses reported by the GDSC and CCLE projects. We received extensive feedback on the comparisons that we performed. This feedback, along with the release of new data, prompted us to revisit our initial analysis. We present a new analysis using these expanded data, where we address the most significant suggestions for improvements on our published analysis - that targeted therapies and broad cytotoxic drugs should have been treated differently in assessing consistency, that consistency of both molecular profiles and drug sensitivity measurements should be compared across cell lines, and that the software analysis tools provided should have been easier to run, particularly as the GDSC and CCLE released additional data. Our re-analysis supports our previous finding that gene expression data are significantly more consistent than drug sensitivity measurements. Using new statistics to assess data consistency allowed identification of two broad effect drugs and three targeted drugs with moderate to good consistency in drug sensitivity data between GDSC and CCLE. For three other targeted drugs, there were not enough sensitive cell lines to assess the consistency of the pharmacological profiles. We found evidence of inconsistencies in pharmacological phenotypes for the remaining eight drugs. Overall, our findings suggest that the drug sensitivity data in GDSC and CCLE continue to present challenges for robust biomarker discovery. This re-analysis provides additional support for the argument that experimental standardization and validation of pharmacogenomic response will be necessary to advance the broad use of large pharmacogenomic screens.
Keywords: cancer; consistency; drug sensitivity; pharmacogenomic agreement; pharmacogenomics.
Conflict of interest statement
Competing interests: No competing interests were disclosed.
Figures







References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

