Comprehensive Evaluation of TFF3 Promoter Hypomethylation and Molecular Biomarker Potential for Prostate Cancer Diagnosis and Prognosis
- PMID: 28930171
- PMCID: PMC5618665
- DOI: 10.3390/ijms18092017
Comprehensive Evaluation of TFF3 Promoter Hypomethylation and Molecular Biomarker Potential for Prostate Cancer Diagnosis and Prognosis
Abstract
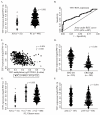
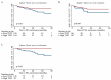
Overdiagnosis and overtreatment of clinically insignificant tumors remains a major problem in prostate cancer (PC) due to suboptimal diagnostic and prognostic tools. Thus, novel biomarkers are urgently needed. In this study, we investigated the biomarker potential of Trefoil factor 3 (TFF3) promoter methylation and RNA expression levels for PC. Initially, by quantitative methylation specific PCR (qMSP) analysis of a large radical prostatectomy (RP) cohort (n = 292), we found that the TFF3 promoter was significantly hypomethylated in PC compared to non-malignant (NM) prostate tissue samples (p < 0.001) with an AUC (area under the curve) of 0.908 by receiver operating characteristics (ROC) curve analysis. Moreover, significant TFF3 promoter hypomethylation (p ≤ 0.010) as well as overexpression (p < 0.001) was found in PC samples from another large independent patient sample set (498 PC vs. 67 NM) analyzed by Illumina 450K DNA methylation arrays and/or RNA sequencing. TFF3 promoter methylation and transcriptional expression levels were inversely correlated, suggesting that epigenetic mechanisms contribute to the regulation of gene activity. Furthermore, low TFF3 expression was significantly associated with high ERG, ETS transcription factor (ERG) expression (p < 0.001), as well as with high Gleason score (p < 0.001), advanced pathological T-stage (p < 0.001), and prostate-specific antigen (PSA) recurrence after RP (p = 0.013; univariate Cox regression analysis). There were no significant associations between TFF3 promoter methylation levels, ERG status, or PSA recurrence in these RP cohorts. In conclusion, our results demonstrated diagnostic biomarker potential of TFF3 promoter hypomethylation for PC as well as prognostic biomarker potential of TFF3 RNA expression. To the best of our knowledge, this is the most comprehensive study of TFF3 promoter methylation and transcriptional expression in PC to date.
Keywords: DNA methylation; TFF3; biomarker; diagnosis; expression; prognosis; prostate cancer.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Haldrup C., Mundbjerg K., Vestergaard E.M., Lamy P., Wild P., Schulz W.A., Arsov C., Visakorpi T., Borre M., Høyer S., et al. DNA methylation signatures for prediction of biochemical recurrence after radical prostatectomy of clinically localized prostate cancer. J. Clin. Oncol. 2013;31:3250–3258. doi: 10.1200/JCO.2012.47.1847. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

