Integrating Enhancer Mechanisms to Establish a Hierarchical Blood Development Program
- PMID: 28930689
- PMCID: PMC5618720
- DOI: 10.1016/j.celrep.2017.08.090
Integrating Enhancer Mechanisms to Establish a Hierarchical Blood Development Program
Abstract
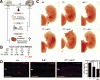
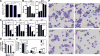
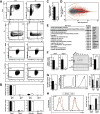
Hematopoietic development requires the transcription factor GATA-2, and GATA-2 mutations cause diverse pathologies, including leukemia. GATA-2-regulated enhancers increase Gata2 expression in hematopoietic stem/progenitor cells and control hematopoiesis. The +9.5-kb enhancer activates transcription in endothelium and hematopoietic stem cells (HSCs), and its deletion abrogates HSC generation. The -77-kb enhancer activates transcription in myeloid progenitors, and its deletion impairs differentiation. Since +9.5-/- embryos are HSC deficient, it was unclear whether the +9.5 functions in progenitors or if GATA-2 expression in progenitors solely requires -77. We further dissected the mechanisms using -77;+9.5 compound heterozygous (CH) mice. The embryonic lethal CH mutation depleted megakaryocyte-erythrocyte progenitors (MEPs). While the +9.5 suffices for HSC generation, the -77 and +9.5 must reside on one allele to induce MEPs. The -77 generated burst-forming unit-erythroid through the induction of GATA-1 and other GATA-2 targets. The enhancer circuits controlled signaling pathways that orchestrate a GATA factor-dependent blood development program.
Keywords: GATA-2; enhancer; erythroid; hematopoiesis; mouse model; myeloid; progenitor.
Copyright © 2017 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




References
-
- Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. - PubMed
-
- Beck D, Thoms JA, Perera D, Schutte J, Unnikrishnan A, Knezevic K, Kinston SJ, Wilson NK, O'Brien TA, Gottgens B, et al. Genome-wide analysis of transcriptional regulators in human HSPCs reveals a densely interconnected network of coding and noncoding genes. Blood. 2013;122:e12–22. - PubMed
-
- Chabot B, Stephenson DA, Chapman VM, Besmer P, Bernstein A. The proto-oncogene c-kit encoding a transmembrane tyrosine kinase receptor maps to the mouse W locus. Nature. 1988;335:88–89. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

